
If you’re familiar with bioluminescence, you’ve probably used it in plate-based experiments to track various biological processes. You understand it provides distinct advantages over traditional fluorescence assays, particularly when it comes to sensitivity. However, there’s always that one nagging question: how representative is the signal on a cell-to-cell level?
Traditional approaches to decipher cell-to-cell signal rely on complex, time-intensive measures that only approximated the findings acquired through bioluminescence. That’s where the GloMax® Galaxy Bioluminescence Imager comes in. This new tool will enhance your ability to visualize proteins using NanoLuc® technology, going beyond simple numeric outputs to reveal what’s happening in individual cells.
NanoLuc® technology is well-known for its ability to assist in detecting subtle protein interactions in complex biological systems. This bright luminescent enzyme enables a much broader linear range than fluorescence, improving detection of small changes in protein activity, such as proteins interacting. Microplate readers measuring NanoLuc® assays rely on signal generated from many cells. This results in an approximation of what is occurring biologically. Truly validating those luminescent readings within a cell population has been challenging—until now. The GloMax® Galaxy allows you to perform bioluminescence imaging, moving beyond the numbers, offering the power to visualize protein interactions directly.
Using NanoBiT® Assays to detect Protein:Protein Interactions
Let’s look at an example of detecting protein:protein interactions through NanoBiT. NanoBiT works through splitting the NanoLuc® Luciferase enzyme into two subunits, Large BiT (LgBiT) and Small BiT (SmBiT). These two proteins have low affinity to each other, and when they come into close proximity LgBiT and SmBiT reconstitute the luciferase enzyme and produce a measurable signal. This non lytic assay provides a way to quantity dynamic protein:protein interactions sensitively in real time.
Researchers use this technology to explore G protein-coupled receptors (GPCRs). They chose CX3CR1 (Chemokine (C-X3-C motif) receptor 1), a highly selective chemokine receptor and surface marker for cytotoxic effector lymphocytes1. Understanding how GPCRs interact with Beta Arrestin 2 (ARRB2) provides a way to better understand mechanisms underlying inflammatory diseases through receptor recruitment.
HEK293 cells stably expressing CX3CR1 fused to SmBiT and ARRB2 fused to LgBiT were used to explore this GPCR. This cell line is available through our Early Access program. These cells were treated with various doses of Fractalkine, an agonist for CX3CR11. Treatment with Fractalkine results in binding to ARRB2 (Arrestin, Beta 2) and receptor recruitment. Figure 2 demonstrates NanoBiT® Luciferase signal generated in a dose-response manner upon treatment with Fractalkine. These results are consistent with prior work published with these same cells2.
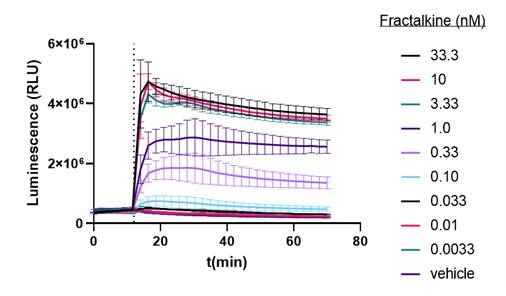
Figure 1: Dose response of Protein:Protein Interaction of CX3CR1 and ARRB2 to Fractalkine
This data is quite useful; however, it does not fully indicate internalization of CX3CR1 upon binding to ARRB2. Bioluminescence imaging was used to confirm the signal indicates receptor internalization.
Transform a Number into an Image Through Bioluminescence Imaging
It is clear that NanoBiT® technology provides a powerful method to detect protein:protein interactions through bioluminescent signals. However, what cannot be gleaned through this plate-based assay is whether intracellular transport of the proteins is generating the bioluminescence. To visualize the signal and internalization, the HEK293 cells were plated at a density of 60-75K cells/well and incubated overnight. The next morning the media was carefully replaced with Opti-MEM and cells were serum-starved for 5 hours before treatment with 10nM of Fractalkine.
Figure 2 shows a video containing images collected on the GloMax® Galaxy. The total capture time was 24 minutes. Each image captures a 3-minute exposure. The beginning of the video shows no cells, indicating no luminescence before treatment with Fractalkine. Treatment with 10nM of Fractalkine leads quite quickly to an increase in luminescent signal, indicative of CX3CR1:AARB2 interaction. A close observation of the video shows localization of the luminescent signal shifting from predominantly the cellular membrane into the intracellular space, appearing more like puncta. This shift in signal over the short time frame indicates receptor internalization after CX3CR1:AARB2 interaction.
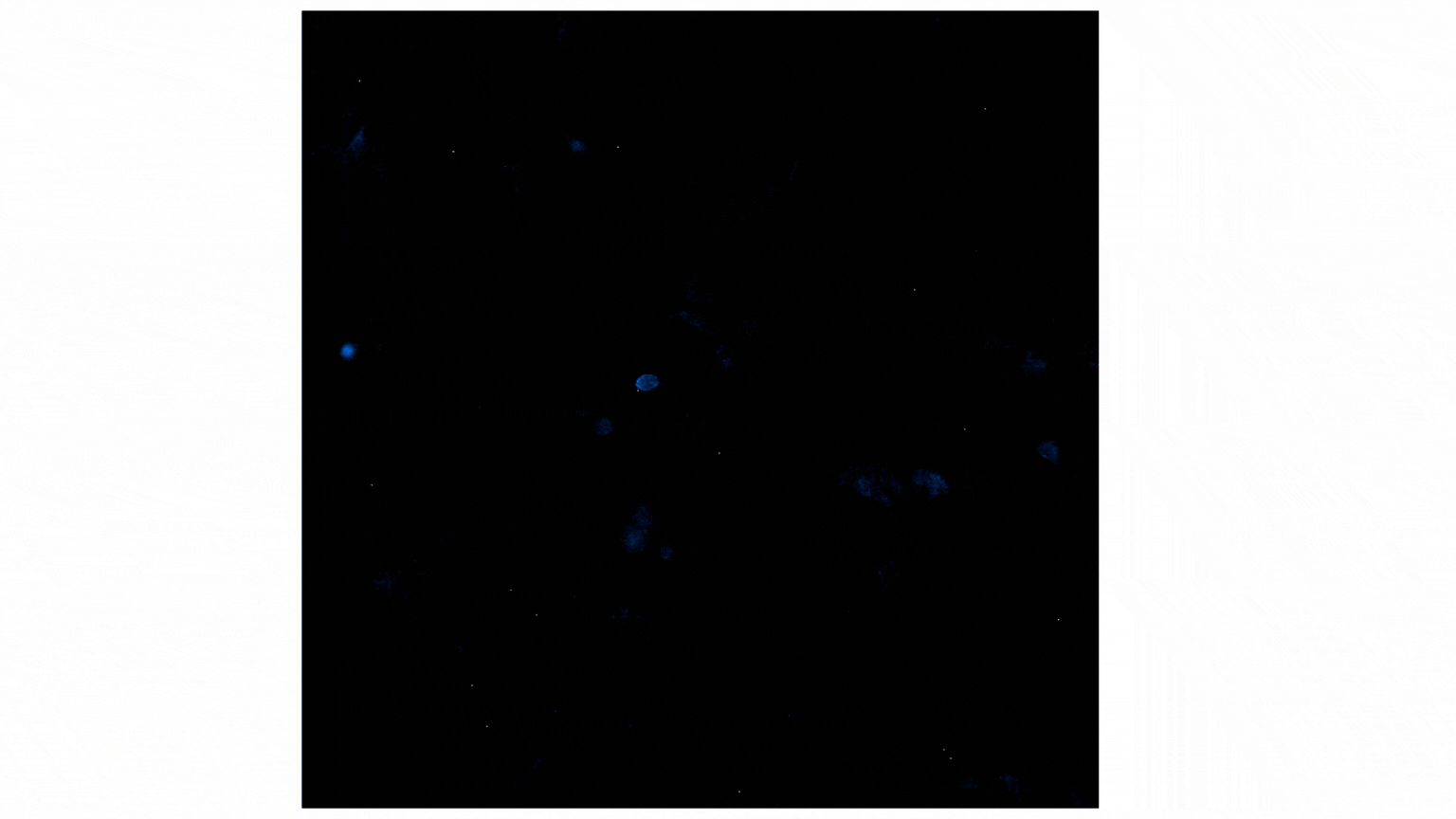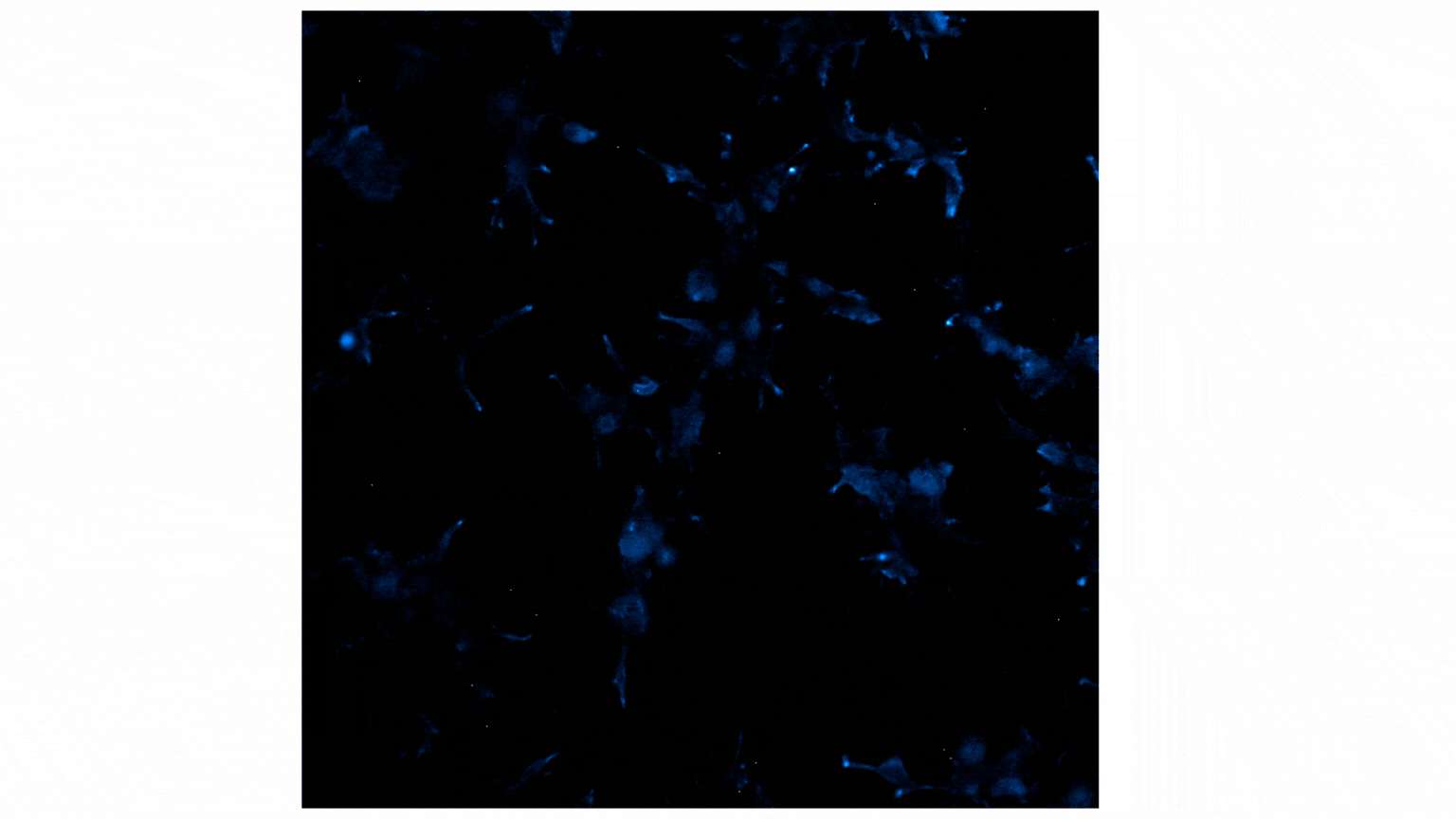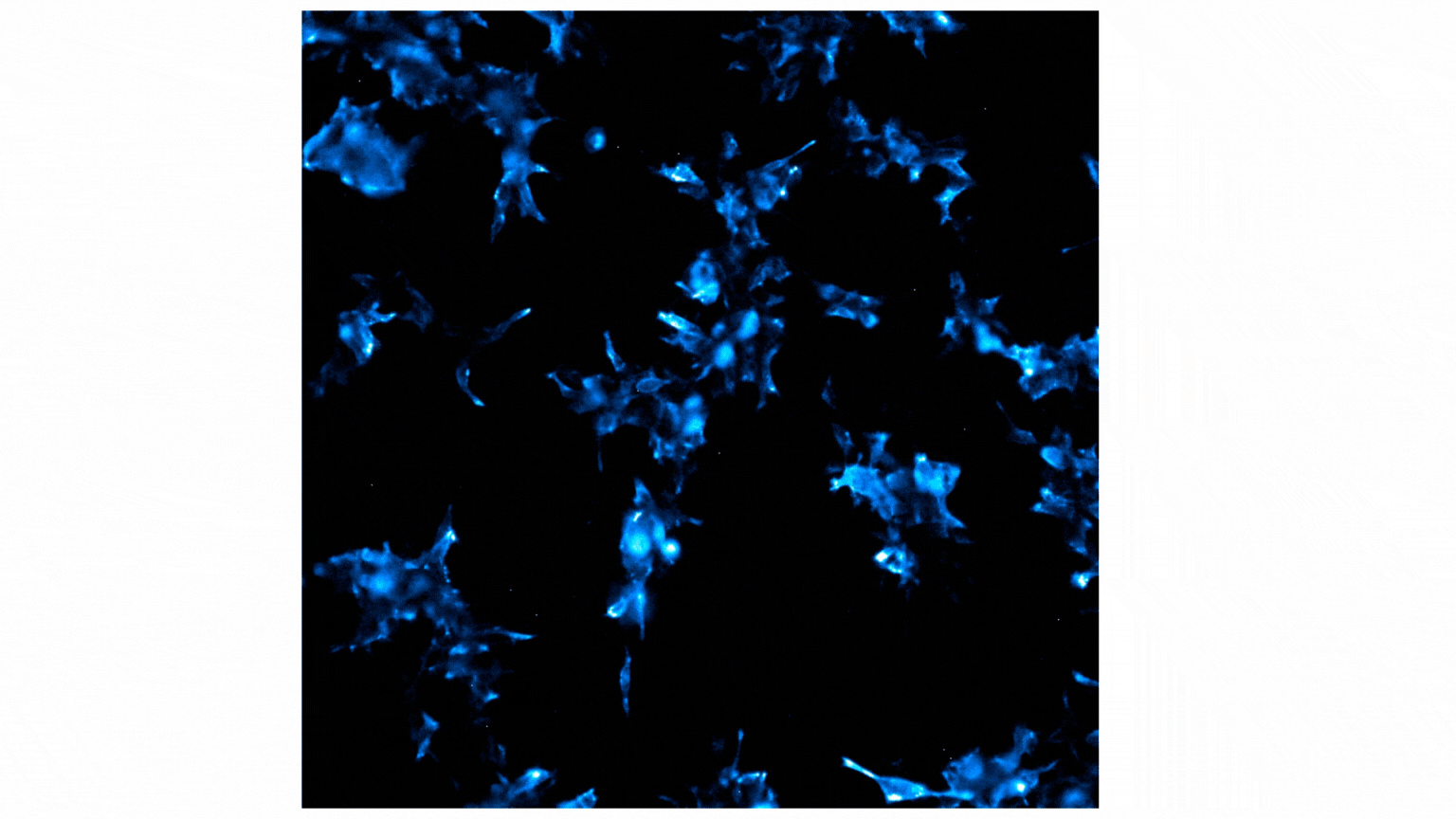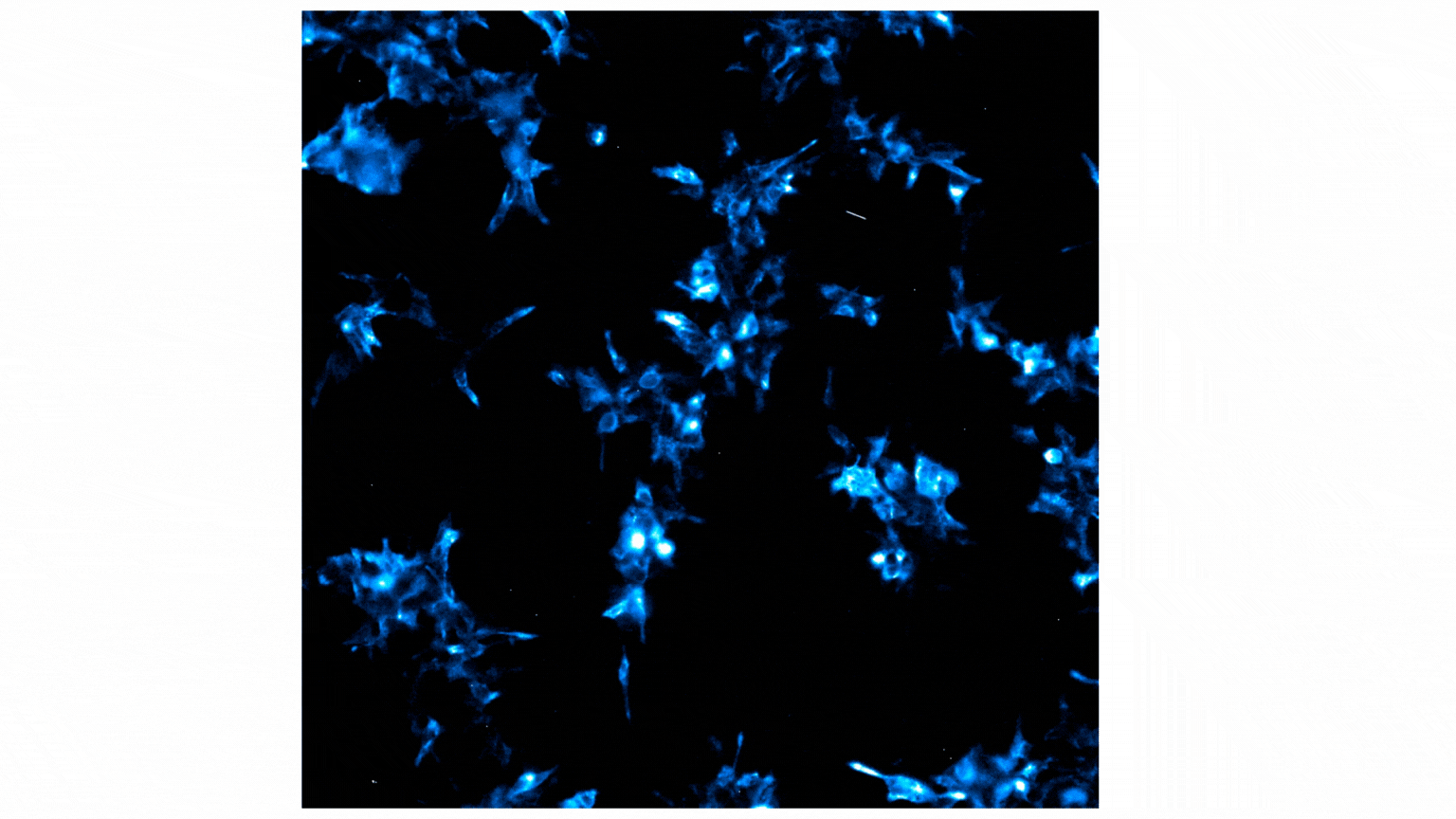
Figure 2: Treatment with Fractalkine induces an increase in NanoBiT® signal over time. Localization shifts from predominantly membrane to intracellular puncta, indicating receptor internalization.
Why is Bioluminescence Imaging Useful?
In practical terms, bioluminescence imaging enables you to finally capture the subtleties in your data. Instead of analyzing signal generated from an entire plate well, the GloMax® Galaxy provides a way to observe signal at the cell-to-cell level. Imaging provides a way to distinguish between responsive and non-responsive cells, reveal how treatments are affecting individual proteins interactions, and helps determine whether the rate of these interactions is consistent within a cell population. By offering real-time insights into biological events, with the GloMax® Galaxy, it’s no longer about just gathering data—it’s about seeing it come to life.
Are you interested in another application using the GloMax® Galaxy? Take a look at this technical article.
Citations
Brescia, P. J., et al. (n.d.). Application Note Drug Discovery and Development A High-Throughput Luminescence-Based Live Cell Assay to Measure β-Arrestin Recruitment in Real Time.
Jones, B. A., et al. (2010). Fractalkine/CX3CL1: A Potential New Target for Inflammatory Diseases (Vol. 10).
