
The use of reporter genes for simple analysis of promoter activity (promoter bashing) is a well known practice. However, there are many other elegant applications of reporter technologies. One such application is illustrated in the paper by Zheng et al., published in the Sept. 2008 issue of Cancer Research. These researchers from the Hormel Institute at the University of Minnesota showed that the cyclin-dependent kinase cdk3 phosphorylates the transcription factor ATF1 and enhances its transcriptional and transactivation activity. The observed cdk/ATF1 signaling was shown to have an important role in cell proliferation and transformation. To do this they used several variations of a reporter-based two-hybrid assay.
These authors used a luciferase-based two-hybrid system (the CheckMate System) to find proteins that interact with cdk3. They first cloned cdk3 downstream of the yeast GAL4 DNA binding domain in the pBIND-cdk plasmid. They also cloned several candidate transcription factors into the interacting pACT-vector, which contains the herpes simplex VP16 activation domain. Both vectors were cotransfected into cells along with the pG5-luc plasmid, which contains five GAL4 binding sites upstream of a firefly luciferase gene. When two cloned proteins interact, the VP16 activation domain and the GAL4 binding domain are brought close enough together to activate transcription of the firefly luciferase gene. Thus, interacting protein partners cause an increase in luciferase expression that is easily measurable.
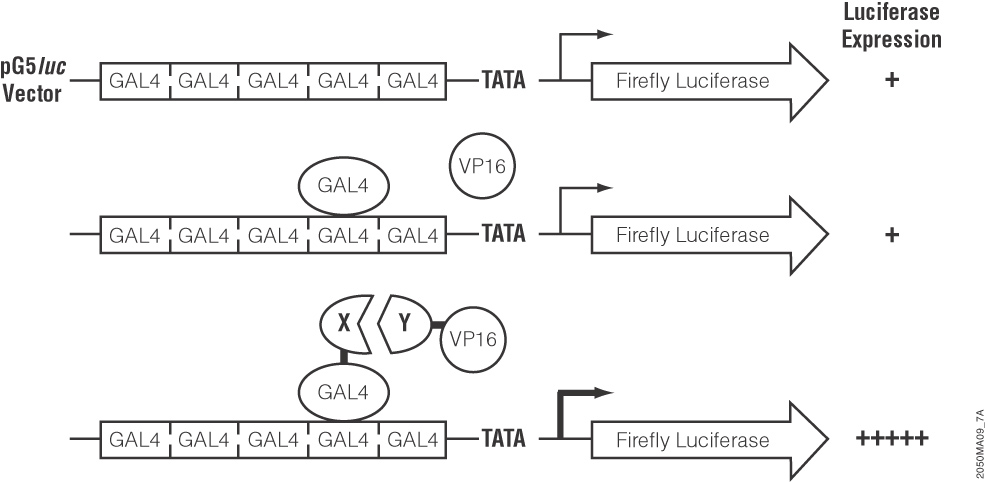
The authors went on to use a creative variant of this type of assay to illustrate that cdk3 phosphorylation of ATF1 was an essential element in EGF mediated transactivation of ATF1. They showed that EGF induced activation of ATF1 was increased in cdk3 expressing cells compared with mock transfected controls, again using luciferase expression as an indicator of active (phosphorylated) ATF1. In that experiment, they cotransfected the plasmid containing the firefly luciferase gene and the upstream GAL4 binding domains along with an expression vector containing cdk3 and a second expression vector containing ATF1 fused to the GAL4 binding domain. After exposure to EGF, the cells were lysed and luciferase activity assayed. Luciferase activity was increased in cdk3 cells compared to mock-transfected cells, showing that cdk3 stimulated EGF-induced ATF1 activation.
Through traditional protein analysis, serine 63 of ATF1 was identified as the phosphorylation target of the cdk3. Replacement of Ser63 with Ala63 (S63A) rendered the ATF1 inactive in the modified CheckMate Assay. Full CheckMate Analysis for the S63A mutant demonstrated that the S63A mutant bound cdk3 just as strongly as the wildtype ATF1 indicating that the Ser 63 was not directly involved with the interaction of the two proteins.
Researchers from the same laboratory used a similar approach in a 2007 paper showing that the kinase RSK2 interacts with NFAT3. In that paper they also used the standard CheckMate assay to identify potential interacting partners of RSK2. They then went on to show that RSK2 induces NFAT activation in a reporter assay where they cloned a minimal interleukin-2 promoter and three NFAT-binding consensus sequences upstream of the luciferase reporter gene. The reporter vector was then cotransfected into 293 cells along with plasmids containing NFAT and RSK2. After exposure to the NFAT inducer A23187, lucferase activity was measured. Luciferase activity was increased with rsk2 in a dose-dependent manner.
This has been a very brief summary of the approach taken in these papers. You can read all the experimental details by following the links below. Also, please use the comments if you would like to share further applications of reporter assays with us and our readers.
Zheng, D. et al. (2008) Cyclin-dependent Kinase 3-Mediated Activating Transcription Factor 1 Phosphorylation Enhances Cell Transformation. Cancer Res. 68, 7650-59.
Cho, Y-Y. et al. (2007) RSK2 Mediates Muscle Cell Differentiation through Regulation of NFAT3. J. Biol. Chem. 285, 8380-92.
More information on use of the original CheckMate Mammalian Two-Hybrid System can be found on our website. We also have a newer system that allows directional cloning using Flexi® Vectors.
Looking for new ways to study protein interactions? This blog describes the use of NanoBRET™ Assays to analyze virus:host protein interactions.
Isobel Maciver
Latest posts by Isobel Maciver (see all)
- 3D Cell Culture Models: Challenges for Cell-Based Assays - August 12, 2021
- Measuring Changing Metabolism in Cancer Cells - May 4, 2021
- A Quick Method for A Tailing PCR Products - July 8, 2019
