Post-translational modifications of proteins are critical for proper protein function. Modifications such as phosphorylation/dephosphorylation can act as switches that activate or inactivate proteins in signaling cascades. The addition of specific sugars to membrane proteins on cells are critical for recognition, interaction with the extracellular matrix and other activities. While we know volumes about some types of protein modifications, ADP-ribosylation on aspartate and glutamate residues has been more difficult to study because of the chemical instability of these ester-linked modifications.
Matić Lab (Eduardo José Longarini and Ivan Matić) recently published a study that explored mono-ADP-ribosylation (ADPr) on aspartate and glutamate residues by the protein PARP1 and its potential reversal by PARG. PARP1 and PARG signaling are central to DNA repair and apoptosis pathways, making them potentially powerful therapeutic targets in cancer or neurodegenerative diseases in which DNA repair processes are often disrupted.
To address the instability of the ester-linked modifications, Matić Lab developed a methodology that preserves ester-linked modifications, allowing the detection of DNA damage-induced mono-ADPr on aspartate/glutamate residues using antibody and mass spectrometry techniques. In their process they adjusted the temperature during cell lysis to room temperature rather than 37°C. Protein samples were never heated above room temperature, and most often samples remained at 4° C. Additionally they reduced the time for protein digestion and conducted it under acidic conditions. To achieve effective digestion at low pH, they used the new Arg-C Ultra, Mass Spec Grade Protease from Promega.
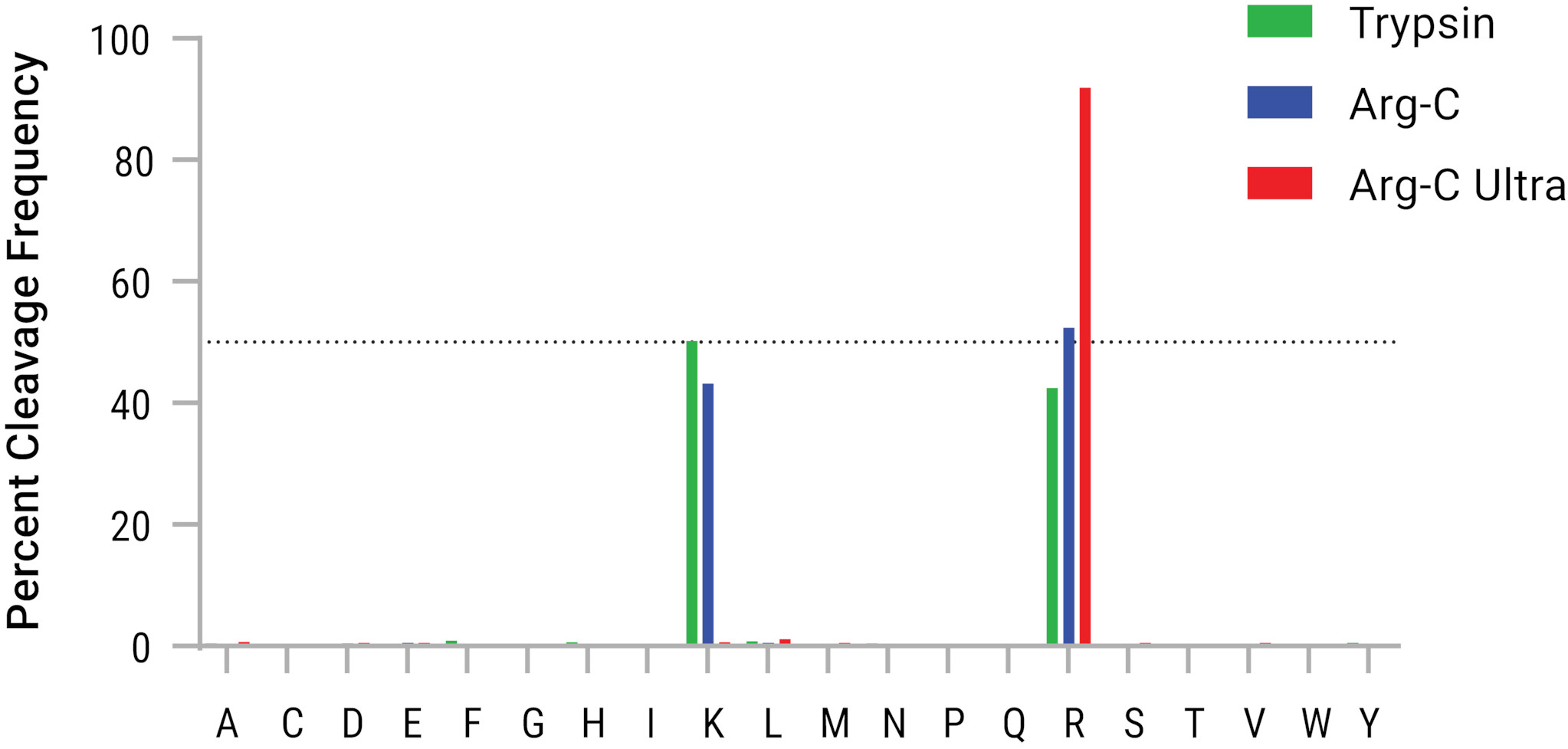
Arg-C Ultra protease enables protein digestion at a mildly acidic pH (around 5.0), which is gentler on ester-linked modifications. Acidic environments are less likely to hydrolyze the ester bonds, allowing the retention of labile ADPr modifications throughout the sample preparation process. Arg-C Ultra protease also provides highly specific and efficient cleavage in a significantly shorter timeframe, reducing time samples are exposed to conditions that would destabilize the ester linkage. In this study, by minimizing degradation, the use of Arg-C Ultra resulted in a roughly 10-fold improvement in the detection of mono-ADPr on aspartate and glutamate residues. This enhanced sensitivity allowed the researchers to identify over 40 high-confidence modification sites, an unprecedented number for this type of PTM.
The authors were able to demonstrate that PARG, traditionally viewed as capable only of breaking poly-ADPr chains, can also reverse ester-linked mono-ADPr in cells. This novel methodology enables further investigation into ADP-ribosylation by various enzymes and broadens the scope for detecting ester-linked modifications, such as ester-linked ubiquitination.
This work provides a refined toolkit for exploring the underappreciated role of ester-linked modifications, enabling insights into how cells respond to damage at the molecular level. Furthermore, understanding these PTMs may enhance the development of PARP inhibitors and related therapies, potentially leading to more targeted and effective treatments for conditions where DNA repair processes are disrupted, such as in cancer and neurodegenerative diseases.
Looking for reliable, high-quality enzymes and surfactants for your mass spec studies? Promega has a host of high-quality mass spectrometry reagents and tools to assist you.
Literature Reviewed
Longarini E.J. and Matić I. (2024) Preserving ester-linked modifications reveals glutamate and aspartate mono-ADP-ribosylation by PARP1 and its reversal by PARG. Nat Commun. 15:4239. DOI:10.1038/s41467-024-48314-0
Michele Arduengo
Latest posts by Michele Arduengo (see all)
- An Unexpected Role for RNA Methylation in Mitosis Leads to New Understanding of Neurodevelopmental Disorders - March 27, 2025
- Unlocking the Secrets of ADP-Ribosylation with Arg-C Ultra Protease, a Key Enzyme for Studying Ester-Linked Protein Modifications - November 13, 2024
- Exploring the Respiratory Virus Landscape: Pre-Pandemic Data and Pandemic Preparedness - October 29, 2024
