Luminescent reporter assays are powerful research tools for a variety of applications. Last March we presented a webinar on this topic, Understanding Luminescent Reporter Assay Design, which proved to enlighten many who registered. The webinar addressed the importance of careful experimental design when using a luminescent reporter such as Promega’s Firefly or NanoLuc® Luciferase.

Reporters provide a highly sensitive, quantifiable metric for cellular events such as gene expression, protein function and signal transduction. Luminescent reporters have become even more valuable for live, real-time measurement of various processes in living cells. This is backed by the fact that a growing number of scientific publications reference the use of the NanoLuc® Luciferase reporter and demonstrate its effectiveness as a reporter assay.
One recent publication in PLoS One showed the utility of bioluminescent binding assays, built around NanoLuc, for analysis of ligand-receptor interactions. In the paper by Ge Song et al., Novel Bioluminescent Binding Assays for Ligand-Receptor Interaction Studies of the Fibroblast Growth Factor Family, the authors demonstrate validation of this binding assay by applying it to the fibroblast growth factor (FGF) family.
The FGF family is a group of small proteins implicated in a number of critical functions including angiogenesis, wound healing and embryonic development. In humans, 22 FGFs have been identified and are widely studied due to their role in a range of developmental and disease processes. FGF2, also known as basic FGF, is a prototype of the FGF family and was first isolated in the early 1970s. Its interaction with FGF receptor 1 (FGFR1) and the mechanism underlying this bond continues to be of great interest.
Song et al. sought to assess the biological activities of both wildtype and mutant FGF2s by assaying their receptor activation potency. Plasmids encoding NanoLuc were provided by Promega and used to construct a serum response element (SRE) controlled NanoLuc
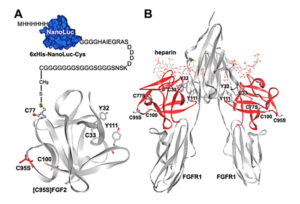
reporter. The reporter was then covalently linked to an appropriate position of FGF2 based on its known tertiary structure. As demonstrated in an earlier research publication, FGF2 folds into a globular structure with four free cysteine residues, two of which are exposed at the surface of the protein. Both are located at the opposite side of the receptor binding site, making them suitable sites for chemical conjugation with the Nanoluc reporter. This engineered reporter construct carries a unique exposed Cys residue at the C-terminus for conjugation with various proteins or peptides (earlier work referenced here).
Key results
Wild-type and mutant versions of human FGF2 were overexpressed, purified and then characterized. The authors tested whether the NanoLuc-conjugated FGF2 could discriminate receptor-binding potencies of different ligands by generating two different versions—each with a tyrosine amino acid (Tyr32 and Tyr111)—and measured how they interacted with FGF receptor.
The biological activities of the recombinant FGF2s were assessed by receptor activation potency and monitored using a serum response element (SRE)-controlled NanoLuc reporter in HEK293T cells expressing endogenous FGF receptor. Both wild-type versions efficiently activated the SRE pathway, consistent with previously reported values of recombinant FGF2. In contrast, the mutant FGF2s carrying a Tyr to Ala mutation demonstrated significantly decreased receptor activation potencies. These results suggested that both Tyr residues are important for FGF2 to activate the endogenous FGF receptor in HEK293T cells.
Next the authors tested the binding of the FGF2-Luc conjugate to FGFR1 by performing saturation binding assays. An overexpressed human FGFR1 transcript was used as the receptor source. FGF2-Luc did bind to living HEK293T cells overexpressing FGFR1 in a typical saturation manner, consistent with previously measured values of radioactive labeled FGF2 tracers. This suggested that the conjugated NanoLuc moiety did not negatively affect FGF2 receptor-binding. Scatchard plot analyses of the specific binding data confirmed one-site binding of FGF-Luc with the overexpressed receptor. The two mutated forms of FGF2 showed significantly decreased binding potency to the receptor.
These results prove that FGF2-Luc can monitor binding of various ligands with the overexpressed receptor FGFR1, representing a novel non-radioactive tracer for ligand-receptor interaction studies.
A saturation binding assay using the endogenous FGF receptor was performed to test the sensitivity of the novel bioluminescent FGF2 tracer. Results demonstrated that the bioluminescent tracer was sensitive enough to detect the endogenous receptor. Both mutant FGF2s showed approximately 100-fold lower binding potency than wild-type FGF2 towards the endogenous receptor, providing further evidence that both Tyr residues are critical for binding of the endogenous FGF receptor.
Application of the assay to other protein/peptide hormones
This study, in conjunction with previous studies by Song and colleagues, demonstrates the value of this novel bioluminescent assay in the functional analyses of many protein/peptide hormones including relaxin, INSL3, chimeric relaxin family peptide R3/I5, ghrelin, leukemia inhibitory factor, erythropoietin and their respective receptors. Now, FGF2 and its receptor can be added to this growing list. What the authors conclude from this impressive body of work is that although the NanoLuc reporter is large, its conjugation does not negatively affect the receptor-binding of a wide range of protein/peptide hormones, provided it designed to attach to an appropriate position in the engineered reporter.
They conclude that their methods of preparing bioluminescent ligands incorporating NanoLuc, both the chemical conjugation method and a genetic fusion method, can be applied to other protein/peptide hormone interaction studies. In summary, NanoLuc-based fusion tracers for novel bioluminescent ligand-receptor binding assays represent a valuable tool.
For more helpful tips and considerations about designing your own luminescent reporter assay, be sure to download the webinar and view two related and in-depth publications on Promega’s PubHub site:
Designing a Bioluminescent Reporter Assay: Choosing Your Experimental Reporter
Designing a Bioluminescent Reporter Assay: Normalization
Nicole Sandler
Latest posts by Nicole Sandler (see all)
- Writing Scientific Papers: Is There More To This Story? - February 3, 2017
- Top Science Books of 2016 - January 4, 2017
- The Role of the NanoLuc® Reporter in Investigating Ligand-Receptor Interactions - December 5, 2016
