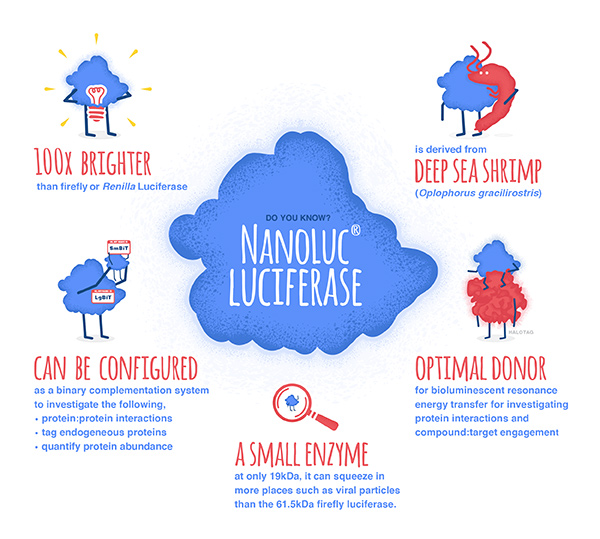
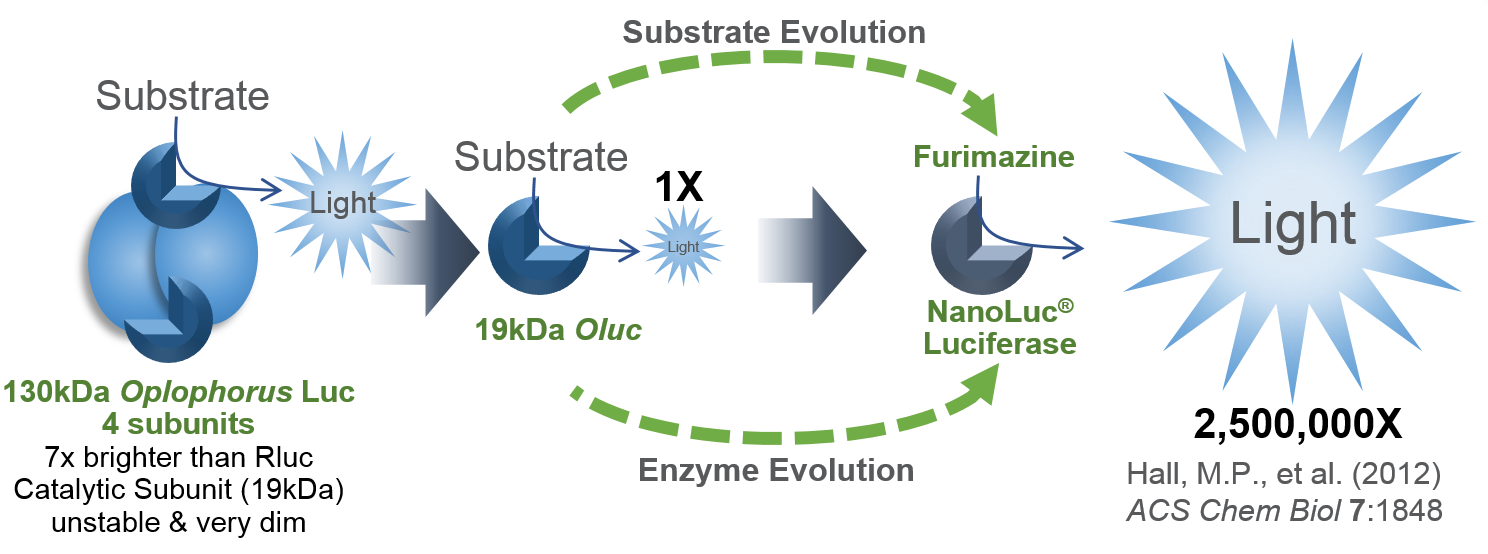
Today’s blog is from guest blogger Ken Doyle of Loquent, LLC. Here, Ken reviews a 2014 paper highlighting specific considerations for using reporter assays to study miRNA-mediated gene regulation.
The accelerated pace of research into noncoding RNAs has revealed multiple regulatory roles for microRNAs (miRNAs). These diminutive noncoding RNA species—typically 20-24 nucleotides in length—are now known to mediate a broad range of biological functions in plants and animals. In humans, miRNAs have been implicated in various aspects of development, differentiation, and metabolism. They are known to regulate an assortment of genes involved in processes from neuronal development to stem cell division. Dysregulation of miRNA expression is associated with many disease states, including neurodegenerative disorders, cardiovascular disease, and cancer.
Typically, miRNAs act as post-transcriptional repressors of gene expression, either by targeted degradation of messenger RNA (mRNA) or by interfering with mRNA translation. Most miRNAs exert these effects by binding to specific sequences called microRNA response elements (MREs). These sequences are found most often within the 3´-untranslated regions (3´-UTRs) of animal genes, while they may occur within coding sequences in plant genes.
Studies of the regulatory roles played by miRNAs often involve cell-based assays that use a reporter gene system, such as luciferase or green fluorescent protein. In a standard assay, the reporter gene is cloned upstream of the 3´-UTR sequence being studied; this construct is then cotransfected with the miRNA into cells in culture. A study by Campos-Melo et al., published in September 2014, examined this experimental approach for miRNAs from spinal cord tissues, using firefly luciferase as the reporter gene and Renilla luciferase as a transfection control.
Continue reading “A Normalization Method for Luciferase Reporter Assays of miRNA-Mediated Regulation” Like this:
Like Loading...