In 1982, picked up because of its homology to chicken virus genes that could transform cells, MYC became one of the first human genes identified that could drive cellular transformation (1,2). Since that time countless laboratories have prodded and poked the human MYC gene, the MYC protein, their homologs in other animal models, and their transforming viral counterparts.
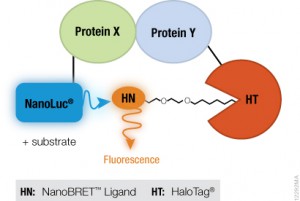
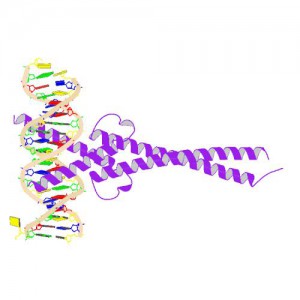
MYC is a transcription factor and forms heterodimers with a required protein partner, MAX, before binding to the E box sequences of DNA regulatory regions (3). MYC regulates gene expression of many targets through interactions with a host of proteins, often referred to as the MYC Interactome (2). In fact, MYC is estimated to bind 10–15% of the genome, and it regulates the expression of genes that are transcribed by by each of the three RNA polymerases (2).
MYC plays a central role in regulating cell growth, proliferation, apoptosis, differentiation and transformation, acting as a central integrator of cellular signals. MYC is tightly regulated at multiple levels from gene expression to protein stability. Dysregulation (usually upregulation) of the amount and stability of Myc protein is observed in many human cancers. Even in cancers in which MYC is not directly involved in transforming cells, its normal expression is often required to support the extracellular matrix and/or vascularization necessary for tumor growth and formation (4).
Because MYC is such a central player cancer pathology, it is an attractive target for cancer therapeutics (2) .
Continue reading “Targeting MYC: The Need to Study Protein:Protein Interactions in Cells”