When I was a post-doc at UT Southwestern, I was fortunate to interact with two Nobel prize winners, Johann Deisenhofer and Fred Gilman. Johann once helped me move a -80°C freezer into his lab when we lost power in my building. I once replaced my boss at small faculty mixer with a guest speaker and had a drink with Fred Gilman and several other faculty members from around the university. Among the faculty, one professor had a cell phone on his belt, an odd sight in 1995. Fred Gilman asked him what it was and why he had it. It was so his lab could notify him of good results anytime of the day. Fred laughed and told him to get rid of it – if it’s good data, it will survive until morning.
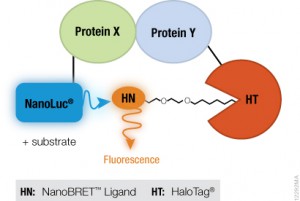
I was reminded of this story when I read a recent paper by Bodle, C.R. et al (1) about the development of a NanoBiT® Complementation Assay (2) to measure interactions of Regulators of G Protein Signaling (RGS) with Gα proteins in cells. (Fred Gilman was the first to isolate a G protein and that led to him being a co-recipient of the Nobel Prize in 1994). The authors created over a dozen NanoBiT Gα:RGS domain pairs and found they could classify different RGS proteins by the speed of the interaction in a cellular context. The interactions were readily reversible with known inhibitors and were suitable for high-throughput screening due to Z’ factors exceeding 0.5. The study paves the way for future work to identify broad spectrum RGS domain:Gα inhibitors and even RGS domain-specific inhibitors. This is the second paper applying NanoBiT Technology to GPCR studies (3).
A Little Background…
A primary function of GPCRs is transmission of extracellular signals across the plasma membrane via coupling with intracellular heterotrimeric G proteins. Upon receptor stimulation, the Gα subunit dissociates from the βγ subunit, initiating the cascade of downstream second messenger pathways that alter transcription (4). The Gα subunits are actually slow GTPases that propagate signals when GTP is bound but shutdown and reassociate with the βγ subunit when GTP is cleaved to GDP. This activation process is known as the GTPase cycle. G proteins are extremely slow GTPases.