MicroRNAs (miRNAs) are small, non-coding RNAs that play a role in regulating cancer by acting as both tumor suppressors and oncogenes. Ranging in size from 18–25 nucleotides, miRNAs function in feedback mechanisms to regulate many cellular processes including cell proliferation, apoptosis, cell signaling and tumorigenesis (1).
Not surprisingly, dysregulation of miRNA expression can have serious repercussions. For example, miRNAs are dysregulated in almost all human cancers (1). Because of the potential to influence cancer growth and development, there is growing interest in miRNA profiling to identify possible biomarkers for cancer diagnosis or prognosis, as well as potential therapeutic targets (1).
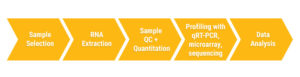
Growing interest in miRNAs as both biomarkers of disease and therapeutic targets drives the need for fast and effective methods for miRNA profiling. Profiling miRNA targets follows a relatively simple workflow: sample selection, RNA extraction, RNA QC and quantitation, RNA profiling and data analysis (2,3). So what happens at each step?