According to the National Human Genome Research Institute, synthetic biology is “a field of science that involves redesigning organisms for useful purposes by engineering them to have new abilities”. Synthetic biology has a broad range of applications, from manufacturing pharmaceuticals and other biologically active chemicals and biofuels, to accelerating the adoption of plant-based burgers (1).
At the heart of the synthetic biology revolution is the rapid technological advancement—and accompanying drop in costs—of DNA oligonucleotide synthesis. Typically, synthetic biology researchers use oligonucleotides as building blocks to assemble genes of interest that are then introduced into, and expressed by, a different organism. For example, to create the plant-based Impossible Burger, the soy leghemoglobin gene (normally found in the root nodules of leguminous plants) was synthesized and expressed in yeast cells (1). This component gives the burger its meaty flavor and appearance of “bleeding” when cooked.

The Synthetic Genome and Minimal Cells
Interest in synthetic biology surged in 2010, when scientists at the J. Craig Venter Institute (JCVI) announced they had assembled a complete, synthetic genome and used it to create a functional bacterial cell (2). The research built upon years of earlier work designing and assembling synthetic genomes in several Mycoplasma species. Combined with optimized techniques that helped propagate the synthetic genome in yeast, these methods helped ensure the genome would remain functional when transplanted into another cell. In the 2010 study, the researchers designed a 1.08Mbp synthetic M. mycoides genome, synthesized its components, assembled a functional genome, and transplanted it into a M. capricolum recipient cell. This new cell was controlled entirely by the synthetic genome, and it was able to replicate continuously to create new, fully functional M. mycoides cells (2).
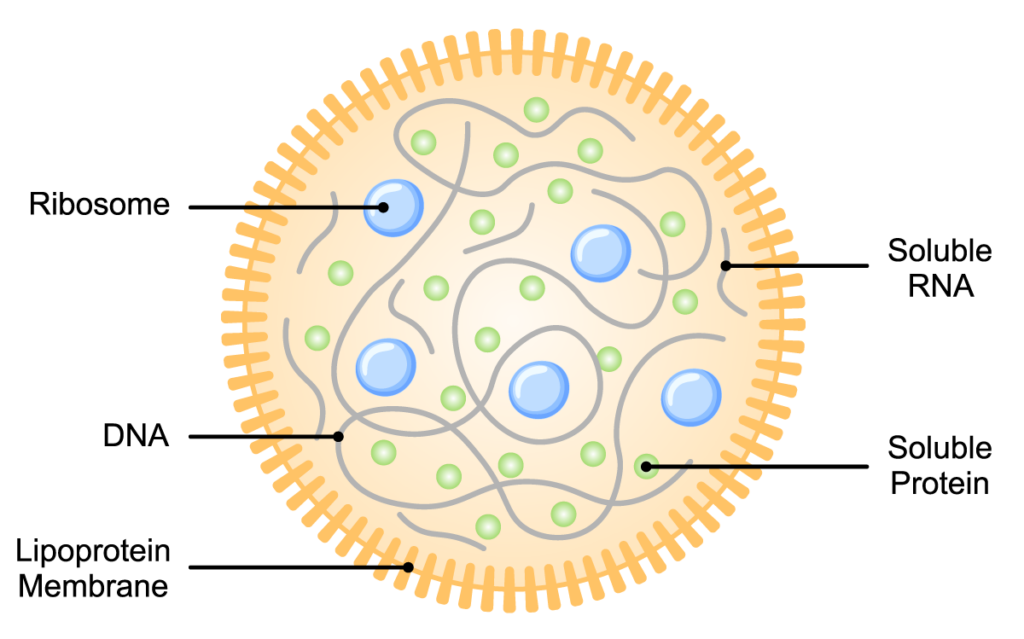
The JCVI research was part of a larger quest—one that had occupied their attention for 15 years prior to the publication by Gibson et al. They focused on Mycoplasma species because the M. genitalium genome, which they had sequenced in 1995, contains the smallest complement of genes of any known organism capable of replicating in laboratory conditions. The researchers wanted to determine which of the 525 M. genitalium genes were essential for growth, so they could design an even smaller, “minimal” genome. In 2016, they announced they had designed and synthesized this minimal bacterial genome (3) which—at 531kbp and containing 473 genes—was smaller than that of any known autonomously replicating cell. The “minimal cells” derived from this minimal genome were able to replicate, although they exhibited some anomalous growth properties. Further refinement of this approach in collaborative studies led to a better definition of the essential metabolic requirements for a minimal cell, resulting in near-normal cell division and morphology (4, 5).
Evolution of a Minimal Cell
In another collaborative study published this month, researchers demonstrated that engineered minimal cells can evolve in response to mutation accumulation experiments (6). Observed mutation rates for the progenitor M. mycoides nonminimal cell and the synthetic minimal cell were the highest among all reported bacteria, but genome minimization did not affect mutation rates. While genome minimization reduced the relative fitness by 50%, almost all of this cost was regained over 2,000 generations of evolution. Further, the minimal cells evolved 39% faster than the nonminimal cells. Therefore, genome minimization did not alter cellular resources in any way that interfered with the minimal cell’s ability to evolve increased fitness. The researchers postulate that genome minimization could even create opportunities for the organism to exploit essential genes through evolution. They conclude that natural selection can rapidly increase the fitness of one of the simplest autonomously growing organisms.
Future developments in synthetic biology could see widespread adoption of substances produced by synthetic cells at larger and more economical scales than current manufacturing processes allow. As the field matures, synthetic organisms of various species could work together to produce food, medicine, and even architectural components for use in space travel (1).
Promega supports young researchers in synthetic biology through the iGEM Grant Program. Learn more about the program and read about the 2023 award winners.
References
- Voigt, C.A. (2020) Synthetic biology 2020–2030: six commercially-available products that are changing our world. Nat. Commun. 11, 6739.
- Gibson, D.G. et al. (2010) Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329 (5987), 52–56.
- Hutchison III, C.A. et al. (2016) Design and synthesis of a minimal bacterial genome. Science 351 (6280), aad6253.
- Breuer, M. et al. (2019) Essential metabolism for a minimal cell. eLife 8, e36842.
- Pelletier, J.F. et al. (2021) Genetic requirements for cell division in a genomically minimal cell. Cell 184, 2430–2440.
- Moger-Resicher, R.Z. et al. (2023) Evolution of a minimal cell. Nature DOI: 10.1038/s41586-023-06288-x
Related Posts
Latest posts by Ken Doyle (see all)
- Will Artificial Intelligence (AI) Transform the Future of Life Science Research? - February 1, 2024
- RAF Inhibitors: Quantifying Drug-Target Occupancy at Active RAS-RAF Complexes in Live Cells - September 5, 2023
- Synthetic Biology: Minimal Cell, Maximal Opportunity - July 25, 2023
