Almost 90% of the human genome is transcribed into RNA, but only 3% is ultimately translated into a protein. Some non-translated RNA is thought to be useless, while some play a significant yet often mysterious role in cancer and other diseases. Despite its abundance and biological significance, RNA is rarely the target of therapeutics.
“We say it’s undruggable, but I would say that ‘not-yet-drugged’ is a better way to put it,” says Amanda Garner, Associate Professor of Medicinal Chemistry at the University of Michigan. “We know that RNA biology is important, but we don’t yet know how to target it.”

Amanda’s lab develops systems to study RNA biology. She employs a variety of approaches to analyze the functions of different RNAs and study their interactions with proteins. Her lab recently published a paper describing a novel method for studying RNA-protein interactions (RPI) in live cells. Amanda says that with the right tools, RPI could become a critical target for drug discovery.
“It’s amazing that current drugs ever work, because they’re all based on really old approaches,” Amanda says. “This isn’t going to be like developing a small molecule kinase inhibitor. It’s a whole new world.”
RNA: A Tricky Target
The central dogma of molecular biology is one of the first things taught in most university-level biology classes. DNA is transcribed into RNA, which is translated into proteins. While the central dogma captures the basic principles of how cells get from nucleic acid to protein, it fails to capture the complex role that RNA plays in human biology. While some RNA such as messenger RNA (mRNA) is translated into proteins, the vast majority is not.
Non-coding RNA (ncRNA) is responsible for regulating transcription, splicing and translation. These molecules include long non-coding RNA (lncRNA), microRNA (miRNA), small interfering RNA (siRNA) and many others. Some types of ncRNA are implicated in many diseases, including metabolic, genetic and neurodegenerative. MicroRNA (miRNA) dysregulation, for example, contributes to cancer growth and progression.
Unfortunately, RNA is notoriously difficult to study. It’s often incredibly small – the miRNAs that Amanda’s lab has been studying are around 22 nucleotides long. RNA easily degrades when isolated, and it’s often susceptible to contamination. In the context of drug discovery, when compared to proteins, it’s much harder to determine the structure of RNA.
“We think of it like a spaghetti noodle changing into many different structures,” Amanda says. “We think of proteins as static – here’s my crystal structure, and I have this beautiful pocket. RNA can’t be crystallized like that because of its flexibility.”
There are some structured RNAs, and Amanda says that cryogenic electron microscopy (Cryo-EM) might elucidate some of those structures. However, even if structured RNAs are discovered, studying them will present new challenges. Biochemical methods that isolate the RNA in solution could fail to recapitulate true biology, due to structural changes or lack of intracellular milieu. To address this challenge, Amanda’s lab needed a way to look at RNA in live cells.
RiPCA: Targeting RNA-Protein Interactions in Live Cells
The RNA interaction with Protein-mediated Complementation Assay, or RiPCA, is a proof-of-concept system used to detect interactions between RBPs and pre-miRNA. The assay uses both HaloTag and NanoBiT and expresses a chemiluminescent signal to indicate an RPI. HaloTag (HT) is a protein fusion tag that forms highly specific covalent bonds with many different ligands. NanoBiT is a two-subunit system based on NanoLuc® Luciferase in which the two subunits, SmBiT and LgBiT, have been optimized for minimal self-association.
RiPCA was designed and tested by two of Amanda’s students, Daniel Lorenz and Sydney Rosenblum. The lab previously worked with an assay called the catalytic enzyme-linked click chemistry assay, or Cat-ELCCA, which combined click chemistry with ELISA-inspired methods to enable high-throughput screening of molecules targeting RPIs. However, Cat-ELCCA was labor-intensive and resource-consuming, and it only looked at RPIs in solution. Daniel proposed a new assay that could examine RPIs in live cells.
“Basically, he’s a chemical biology grad student who knows every cool piece of chemical biotechnology,” Amanda says of Daniel. “He’s like a mad scientist, asking ‘What if we piece all these things together?’ When he came up with this assay, I saw it was pretty complex with a lot of transfection. All these things have to come together for it to work. But I said ‘Let’s try it.’”
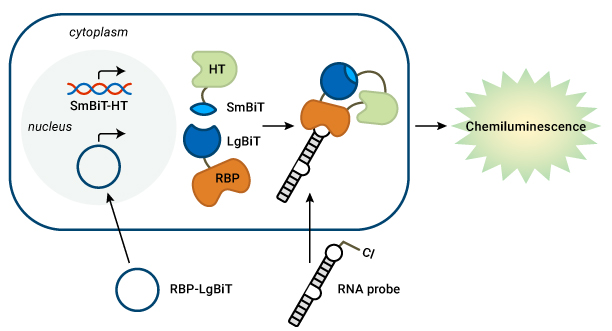
The RiPCA system starts with a cell line expressing SmBiT-HT. The cells are next transfected with a plasmid containing the RBP fused with LgBiT, as well as an RNA probe containing a PEGylated chloroalkane motif. In the cell, the chloroalkane handle first binds to HaloTag. If the RNA probe then binds with the RBP, SmBiT and LgBiT will be brought together to reassemble a functional NanoLuc® Luciferase protein. The cells are then treated with a luciferase substrate, and a luminescent signal indicates successful interaction between the RNA and RBP. In their recent article in RSC Chemical Biology, Amanda’s lab showed that RiPCA can selectively detect the interaction between the pre-miRNA pre-let-7 with its binding partner, Lin28.
“We’re now simultaneously applying it to many other systems, including RNAs outside of miRNAs,” Amanda says. “And we just miniaturized it to 384-wells. We’re gearing up to do a small molecule screen to actually see if this will yield bioactive targeted compounds against let-7 and Lin28.”
Targeting RNA in Drug Development
The Garner Lab isn’t just interested in discovering interactions between RNAs and RBPs. Amanda says their biggest goal is to find RPIs that can be useful for therapeutics research.
“Ultimately, we want to find a molecule that has some kind of functional effect. We don’t really care if it targets the RNA or the RNA-binding protein, because we’ve purposely taken this broader view. Maybe let-7 doesn’t have enough structural complexity that we’ll ever get a very specific compound. Maybe going after the RNA-binding protein is better. And there may be RPIs that we can’t target with a small molecule, but that doesn’t mean that it couldn’t be targeted with a biologic.”
By taking a broad approach with diverse methods, Amanda hopes that her lab can open new frontiers in RNA research. She encourages other labs to use their assays, including the RiPCA system, and she hopes to see her research used in biotechnology and pharmaceuticals.
“There aren’t a lot of validated RNA-binding protein interactions in the literature. I think as more and more work goes into this, having an assay that has been demonstrated to be applicable to many systems will hopefully enable this whole new field of going after these RNA-protein interactions,” she says.
Amanda admits that the breadth of her lab’s research and approaches can be overwhelming for new students who aren’t expecting to see RNA analysis, proteomics, and high-throughput screening in the same space. However, she believes that an open mind and a focus on the future are crucial for answering the myriad questions that remain open in her field.
“Some days you’re just like ‘Oh God, why did we get into all of these things?’” she says. “But that’s what science is to me. It’s all about discovery and thinking about the next problems. We read our molecular biology textbook and think we have it figured out, but the more we apply new technologies and modern approaches, the more we realize it might not really happen that way.”
Check out the full methods for detecting RNA-protein interactions in live cells with HaloTag® and NanoBiT® in Amanda’s recent publication in RSC Chemical Biology:
Rosenblum, S.L. et al. (2021) A live-cell assay for the detection of pre-microRNA-protein interactions. RSC Chemical Biology 2, 241-7.
NanoLuc® Luciferase and HaloTag® Technology have a wide range of applications beyond studying RNA-protein interactions. Learn more in these recent blog posts:
- Oh, The Ways You Can “Glo” – Technical Services Scientist Joliene Lindholm gives an overview of how luminescence could support your research.
- A Closer Look at C. difficile Biology with Luminescent Tagging – A recent study used a form of the NanoBiT® enzyme to examine chromatin-binding proteins in C. dificile.
- Choosing a Tag for your Protein – A quick guide to deciding which protein tag is best suited for your research.
