On September 25, Promega Research Scientist David Mokry addressed a full audience at the International Symposium on Human Identification. The event brings together people from the forensic DNA industry – criminalists, analysts, lab directors and more – eager to learn about advancements in the field. Over the next 20 minutes, David unveiled a novel enzyme designed to tackle a challenge that has plagued DNA forensics for decades.
Known as “Reduced Stutter Polymerase,” the new enzyme virtually eliminates confounding stutter artifacts in forensic DNA analysis. When incorporated into STR analysis kits, it will dramatically simplify mixed sample deconvolution and help forensic analysts generate accurate profiles of multiple contributors. This technology is the result of years of collaboration between the Genetic Identity R&D Group and the Advanced Technology Group at Promega.
Here’s how they did it, and why it’s so important.
What is Stutter?
Forensic DNA analysis is based on the amplification and detection of short tandem repeats, or STRs. STRs are DNA segments where short sequences of bases are repeated multiple times. The number of repeats at each locus varies among individuals, allowing analysts to create a unique genetic profile by analyzing several loci.
Polymerase chain reaction, or PCR, is used to amplify each specific STR locus for analysis. During PCR, Taq polymerase sometimes “slips” on the DNA template, resulting in stutter – a shorter DNA product with one or more fewer repeats than the original.
When the sample is separated by capillary electrophoresis, the electropherogram will peaks representing the number of repeats at each locus in the sample. Due to polymerase slippage, small “stutter peaks” may also appear, representing one or more repeats shorter (or occasionally longer) than the actual DNA sequence.
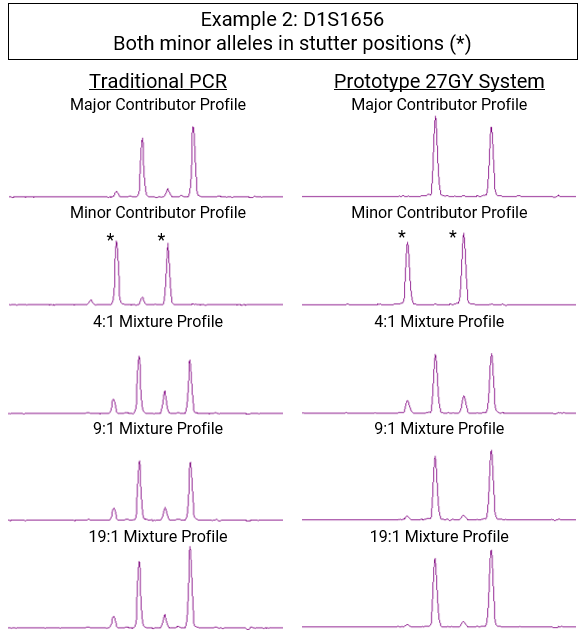
Stutter is not usually an issue with DNA from a single known individual. An analyst can easily filter out the small stutter peaks from the large allele peaks. However, in mixed samples, such as a swab from a public surface, DNA from multiple contributors may be present. In these cases, distinguishing true alleles from stutter artifacts becomes critical. Forensic DNA analysts must sort through these small peaks to filter out those caused by stutter. This process is challenging, time-consuming and sometimes ambiguous, yet essential for accurate criminal casework.
Addressing the Problem

Promega is not the first to try and solve this problem. For nearly four decades, scientists have tried many strategies to eliminate stutter in STR analysis – changing buffers, adjusting concentrations, modifying protocols and much more. Despite these efforts, reliable elimination of stutter remained elusive, leading many to believe it was an unavoidable artifact of STR analysis.
When Promega researchers took on this challenge several years ago, they focused on the enzyme itself. They questioned whether it might be possible to engineer a polymerase that does not slip like Taq polymerase. Their approach involved two main stages of enzyme design.
First, the team examined the protein structure of Taq and incorporated a segment from a polymerase found in T7, a bacteriophage that infects E. coli. This piece, called the Thioredoxin-Binding Domain (TBD), binds a protein called thioredoxin, which increases T7 DNA Polymerase’s affinity for the template DNA strand.
The Enzyme Design team synthesized many iterations of their construct and passed each one to Nick Courtney in the Genetic Identity group to run STR samples and analyze the results. For a long time, their attempts failed to notably reduce stutter. Then one day, just to try something wild, they decided to flood the sample with a huge surplus of thioredoxin – way more than they had ever added before. Nick ran the samples overnight on the Spectrum CE System. When he looked at the data the next morning, he was shocked. They had achieved an 85% reduction in stutter.
Pushing it Further
Achieving an 85% reduction was a groundbreaking result, but the team wanted to push the limits of what they could accomplish. They engineered the thioredoxin into the Taq-TBD construct that eliminated the need for adding excess thioredoxin to the sample. Then, to further optimize the enzyme, they employed a machine learning model to refine the amino acid sequence of the polymerase construct.
This program works the same way as the predictive text algorithm in your phone’s messaging app. When presented with a sequence of words, such as “Once upon a ______,” the predictive text algorithm will analyze everything it knows to find the words most likely to come next. In this case, “time” may be the logical choice, but poetry readers may have a strong case for “midnight dreary.”
When working with proteins, instead of predicting words, the model predicts amino acids. Trained on millions of known protein sequences, the model determines which amino acid is most likely to come next in a sequence to achieve the desired effect – in this case, reducing slippage and stutter.
The team put their sequence through the model, tested various mutations, and combined those that further reduced stutter.
The end result was a tenfold reduction in stutter artifacts, making them undetectable against baseline instrument noise.
In short, stutter is gone.
Why does this matter?
David’s presentation blew away the crowd at ISHI. In a feedback survey, attendees described the massive time savings and data quality improvements this technology would bring to forensic labs.
Beyond the lab, this technology holds major implications for the courtroom. In legal cases, stutter introduces a margin of uncertainty in DNA evidence, which can be exploited by either side. BY virtually eliminating stutter, DNA evidence becomes more straightforward and reliable, reducing the risk of wrongful convictions or acquittals.
To learn more about this technology, watch our video interview with Genetic Identity R&D Director Bob McLaren.
Reduced Stutter Polymerase was also featured in this recent article in The Scientist.
Latest posts by Jordan Villanueva (see all)
- Tackling Undrugged Proteins with the Promega Academic Access Program - March 4, 2025
- Academic Access to Cutting-Edge Tools Fuels Macular Degeneration Discovery - December 3, 2024
- Novel Promega Enzyme Tackles Biggest Challenge in DNA Forensics - November 7, 2024
