Sunscreen usage is increasing, with more people using SPF to prevent the very real threats of skin cancer and early signs of aging. While slathering on the sunscreen is unarguably important to protect your skin from the sun, new concerns arise linking sunscreen chemicals to coral reef bleaching, as an estimated “14,000 tons of sunscreen is believed to be deposited in the oceans annually.”
Coral reefs are the most productive marine ecosystem known. Coral reefs protect coastlines from storm surge and support commercial and recreational fisheries and tourism. Unfortunately, certain chemicals in sunscreen are causing coral reefs to bleach; thus, becoming more susceptible to viral infections. The reefs eventually turn white and die. Coral reef bleaching is the leading cause of coral reef deaths worldwide. This conversation is an important one to discuss leading up to the celebration of World Oceans Day on June 8.
Chemical recreational sunscreen contains oxybenzone, a toxic synthetic molecule. Oxybenzone is prevalent in the majority of mainstream sunscreen brands. This ingredient results in extreme harm to marine organisms. The Ocean Foundation emphasized that, “A single drop of this compound in more than 4 million gallons of water is enough to endanger organisms.” Even if you do not physically go in the water, the chemical can be washed into the ocean through the sand.
In response to this issue, many countries and resorts are banning “reef-toxic” sunscreen. Hawaii and Key West recently passed a bill banning the sale and distribution of any sunscreen that contains 10 toxic ingredients, including oxybenzone. This bill goes into effect January 2021. Many dermatologists are concerned for public safety, highlighting that banning certain sunscreens will decrease overall use. Unprotected sun exposure it the most preventable risk factor for skin cancer. From the perspective of a customer, it is important to be actively informed on what constitutes a “reef-safe” sunscreen. Oxybenzone can pop-up in many moisturizers, primers, and foundations that contain SPF. Reef-friendly options include: any version of chemical sunscreen that does not contain oxybenzone.
With a commitment to protect the environment, Promega has pledged $3 million over the next three years to the Revive and Restore Catalyst Science fund. Organization founders and scientists are focused on an extremely long-term view of wildlife conservation. This fund invests in proof-of-concept research projects that offer innovative solutions for conservation challenges and threatened ecosystems. Marine biologist Steve Palumbi was awarded the first Fund grant to investigate the triggers that may cause corals to bleach. Palumbi reflects on his research in an interview with Stanford News stating, “The report reflects a sense of urgency. We need to start helping corals now, so that as the climate gets worse—and it will inevitably get worse—we’re a little bit in front of the problem. There’s this amazing sense that we all have to just jump in and try ideas and fail so that, eventually, someone comes up with the answers we need.”
Like this:
Like Loading...


 No protein is an island. Within a cell, protein-protein interactions (PPIs) are involved in highly regulated and specific pathways that control gene expression and cell signaling. The disruption of PPIs can lead to a variety of disease states, including cancer.
No protein is an island. Within a cell, protein-protein interactions (PPIs) are involved in highly regulated and specific pathways that control gene expression and cell signaling. The disruption of PPIs can lead to a variety of disease states, including cancer.
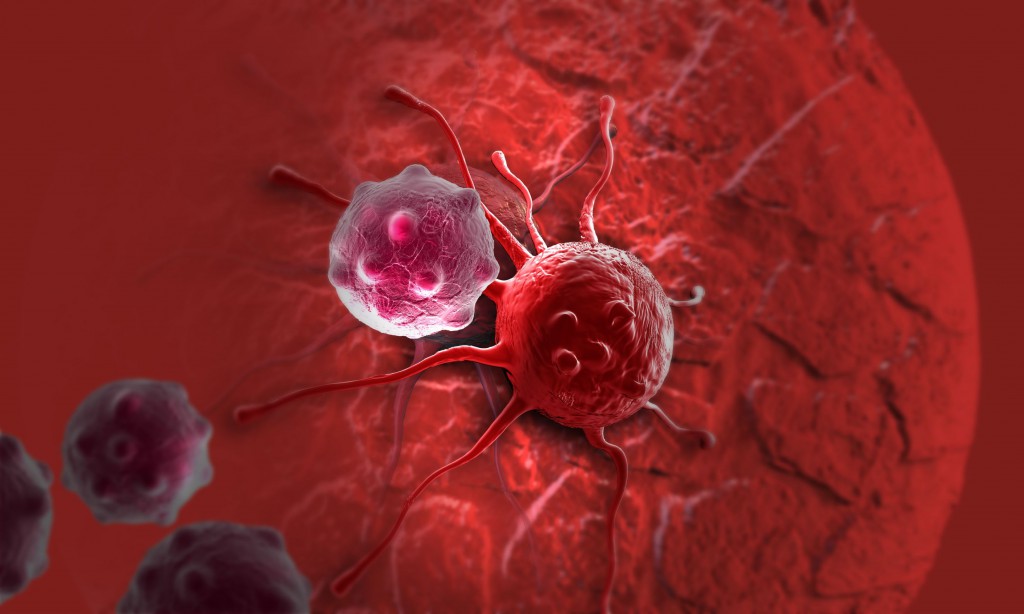 In recent years, great advances have been made in the field of immunotherapy to treat cancer. One of the most promising treatments involves engineering immune cells to express chimeric antigen receptors (CAR). These receptors are carefully designed to recognize antigens expressed on the surface of tumor cells. Once the target is recognized, the CAR-engineered immune cells can attack and kill the tumor cells. CAR T cells have been successfully used to treat certain blood cancers—three CAR T therapies for lymphoma and leukemia have gained US FDA approval. In these cases, T cells were taken from individual patients, grown and genetically-altered in the lab, then reintroduced into the same patient.
In recent years, great advances have been made in the field of immunotherapy to treat cancer. One of the most promising treatments involves engineering immune cells to express chimeric antigen receptors (CAR). These receptors are carefully designed to recognize antigens expressed on the surface of tumor cells. Once the target is recognized, the CAR-engineered immune cells can attack and kill the tumor cells. CAR T cells have been successfully used to treat certain blood cancers—three CAR T therapies for lymphoma and leukemia have gained US FDA approval. In these cases, T cells were taken from individual patients, grown and genetically-altered in the lab, then reintroduced into the same patient. 
