Introduction
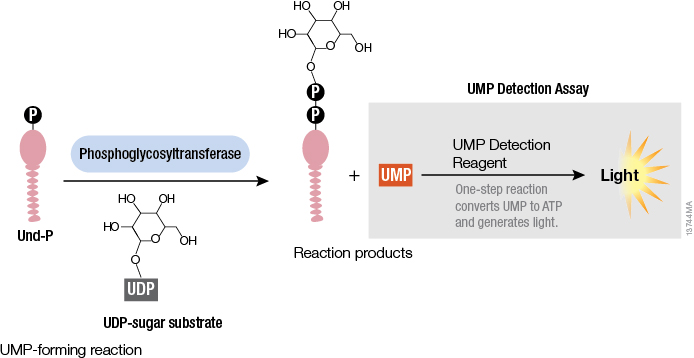
Studying cellular molecules can be challenging. Some processes are troublesome to study due to the lack of an assay or a complicated assay exists but lacks sensitivity. Membrane proteins in particular are difficult to isolate and characterize. Phosphoglycosyltransferases (PGTs) are transmembrane proteins that transfer phosphosugars onto phospholipids, initiating the synthesis of oligosaccharides in bacterial cell walls. This transfer creates a diphosphate link between a lipid and a sugar and generates UMP as a byproduct. Once this lipid–P–P–sugar linkage occurs, more sugars can be added by glycosyltransferases, generating membrane-based polysaccharides (e.g., peptidoglycan) used for signaling, recognition and defense.
While PGTs have been studied biochemically and an X-ray structure of one member exists, much is still unknown about these enzymes. Overexpressing and purifying membrane proteins remains a challenge, and the conventional PGT assay requires isotope labeled-UDP-sugar donors and is based on the solubility difference between substrate and product to determine enzyme turnover using extraction-based or chromatographic methods. While there are other assays that use fluorescent modified substrates or multienzyme analysis, none of the methods can be applied to all of the diverse PGT enzymes.
All PGTs generate UMP as a byproduct of the transfer of a phosphosugar to a phospholipid. Based on the principle of the luminescent UDP-Glo™ Glycosyltransferase Assay where UDP released during the glycosyltransferase reaction was quantitated, a new luminescent assay called UMP-Glo™ Assay is able to measure the activity of PGT enzymes by adding a single reagent to detect UMP. Das et al. validated this assay by testing PglC, a PGT from Campylobacter jejuni, as well as PglC from Helicobacter pullorum and WecA from Thermatoga maritime and published the results in Scientific Reports.
Assay Linearity
To determine the linearity of the UMP-Glo™ Assay, a standard curve of 62.5nM–8μM UMP was prepared and mixed with the UMP Detection Reagent. The luminescence measured from the assay was linear over the UMP concentrations tested. Using PglC from C. jejuni, PGT activity was initiated by mixing the enzyme with its two substrates. The UMP Detection Reagent was added at 0, 5, 10, 15 and 20 minutes, showing UMP-Glo™ Assay linearity over time. In addition, if any of the PglC reaction components were missing, minimal luminescence was measured, indicating little if any UMP was released by the PGT reaction. A radioactive assay with the same enzyme and substrates showed a similar rate and linearity as the UMP-Glo™ Assay, reinforcing the ability of the bioluminescent assay to measure PGT enzyme kinetics.
Detergent and Solvent Tolerance
Because detergents are needed when purifying membrane proteins, the UMP-Glo™ Assay would need to tolerate the presence of detergents with minimal effect on signal intensity. For both Triton X-100 and n-dodecyl β-D-maltoside (DDM), 1% of either detergent had no effect on the UMP Detection Reagent. DMSO is used to solubilize PGT reaction components and inhibitors. Therefore, the UMP-Glo™ Assay signal needs to remain unaffected in the presence of DMSO, and Das et al. confirm that 10% DMSO has no effect on luminescence.
Assay Kinetics
To study the kinetics of C. jejuni PglC, the enzyme was expressed and purified from E. coli and added to two reactions: One with the substrate undecaprenol phosphate (Und-P) at the same concentration while varying the concentration of the substrate UDP-diNAcBac, and one where UDP-diNAcBac concentration remained static and Und-P concentration was varied. The kcat values were similar for both reactions and the Km determined for UDP-diNAcBac was 24.61 while the Km measured for Und-P was 7.18.
Substrate Testing
The substrates for C. jejuni PglC were known, but the UDP-sugar substrate of PglC from H. pullorum has yet to be determined. Previous mass spectrometry experiments suggested that this enzyme used a UDP-HexNAc substrate so Das et al. investigated if UDP-GlcNAc could be used as a substrate. Performing a time course experiment using H. pullorum PglC with UDP-GlcNAc and Und-P showed a similar linear signal over 20 minutes but at a slower rate compared to C. jejuni PglC and its substrates. Thus, the activity of both C. jejuni PglC and H. pullorum PglC could be measured using the UMP-Glo™ Assay.
Enzyme Preparation
Not surprisingly, two enzymes that share the same name from two different organisms have similarity in structure and kinetics. However, WecA from T. maritime is a PGT with different topology from either PglC enzyme, including 11 predicted transmembrane helical domains (TMHDs). Unfortunately, purifying a protein with TMHDs is problematic. When WecA was isolated in the cell envelope fraction (CEF), there was high background in the UMP-Glo™ Assay even in the absence of substrates. WecA with a polyhistidine tag was partially purified on a nickel column despite poor binding, and the resulting preparation was used in the UMP-Glo™ Assay. While the WecA reaction needed more time to produce UMP, the UMP production was linear for 10–40 minutes. WecA activity was confirmed by removing each of the reaction components and demonstrating that there was little UMP produced without both substrates and the enzyme preparation. In addition, increasing amounts of the substrate UDP-GlcNAc in the WecA reaction also showed increasing amounts of UMP produced. These results indicate WecA activity was successfully measured using the UMP-Glo™ Assay.
Screening Capabilities
Because PGTs play a role in bacterial glycan synthesis, these enzymes are important targets for compounds. The UMP-Glo™ Assay can be performed in multiwell plates, making it amenable for high-throughput screening of potential inhibitors. While other published results by the same researchers showed how well the UMP-Glo™ Assay works when screening uridine analogs to inhibit PglC, they discovered that smaller uridine fragments inhibited the luminescent UMP detection assay. Das et al. showed even 5µM uridine was enough to significantly reduce the luminescent signal; 50µM completely inhibited the UMP-Glo™ Assay. Because of this observation, researchers recommended performing two plates in parallel, one with PGT reaction plus compound and one with the potential inhibitor only, to determine if there is any inhibition of the UMP-Glo™ Assay.
Summary
Using the luminescence-based UMP-Glo™ Assay, Das et al. were able to successfully measure the activity of three different PGTs from three different bacteria. By measuring UMP, the byproduct of PGTs, based on the light generated, the UMP-Glo™ Assay can be used to identify UDP-sugar substrates and polyprenols for a wide array of PGT enzymes. Furthermore, the UMP-Glo™ Assay can be used for high-throughput screening, making it easier to test compounds that may inhibit PGTs in bacteria.
Update: The bioluminescent assay described in the referenced article to measure UMP and CMP, byproducts of phosphoglycosyltransferase reactions, is now available and can be ordered as the UMP/CMP-Glo™ Glycosyltransferase Assay.
Reference
Das, D., Walvoort, M.T., Lukose, V. and Imperiali, B. (2016) A rapid and efficient luminescence-based method for assaying phosphoglycosyltransferase enzymes. Sci Rep. 6, 33412. doi: 10.1038/srep33412.
Sara Klink
Latest posts by Sara Klink (see all)
- A One-Two Punch to Knock Out HIV - September 28, 2021
- Toxicity Studies in Organoid Models: Developing an Alternative to Animal Testing - June 10, 2021
- Herd Immunity: What the Flock Are You Talking About? - May 10, 2021
