Updated November 21, 2023
In the life of a cell, phosphorylation of proteins is an everyday occurrence. The transfer of a phosphate group, from a molecule such as adenosine triphosphate (ATP) to a specific functional group on a protein, is catalyzed by a protein kinase. The vast majority of protein kinases are classified as either serine/threonine kinases or tyrosine kinases; over 500 kinase genes have been identified in the human genome (1).
Protein phosphorylation is a key step in most cell signaling pathways, in response to external or internal stimuli, and it is not surprising that dysregulation of these pathways contributes to a variety of cancers. The first oncogene to be characterized was SRC, a gene that encodes a tyrosine kinase (reviewed in 2). With more kinases being implicated in oncogenic pathways, significant drug discovery efforts have been devoted to developing and characterizing inhibitors of protein kinases. These efforts have accelerated ever since the first targeted small-molecule kinase inhibitor, imatinib, received US FDA approval in 2001 for the treatment of chronic myeloid leukemia (3). Since that time, many more protein kinase inhibitors have received FDA approval, with 67 small-molecule inhibitors listed as of September 2021.
In order to be effective, a kinase inhibitor must inhibit a specific kinase target with minimal or no effects on other related kinases. Typically, several small-molecule drug candidates are designed and synthesized to target a specific kinase; each compound is then tested or “profiled” against a large number of related kinases to establish its specificity and selectivity. Ideally, this profiling would be performed in live cells to obtain physiologically relevant data. Traditionally, profiling experiments have used biophysical methods to study the interaction between a compound of interest and its target kinase (target engagement); however, highly sensitive, high-throughput technologies based on luminescence energy transfer are growing in popularity (4).
A Better BRET
Bioluminescence resonance energy transfer (BRET) technology, like fluorescence resonance energy transfer (FRET), is a proximity-based assay in which energy is transferred from a donor to an acceptor when the two are in close proximity. The method was originally developed to study protein-protein interactions in bacteria (5). Unlike FRET, BRET does not require an external light source, because the donor is a luciferase enzyme that generates light through a chemical reaction. Early BRET methods used Renilla luciferase (36kDa) as the donor (6).
In 2014, Promega introduced NanoBRET™ technology that used the small (19kDa), bright NanoLuc® Luciferase as the donor (7,8). The technology achieved substantial improvements in sensitivity and dynamic range over conventional BRET methods, due to greater spectral separation between the donor and acceptor, as well as higher donor light intensity.
NanoBRET™ Kinase Target Engagement
The award-winning NanoBRET™ technology has emerged as a popular method for studying kinase target engagement (TE). Typically, the kinase of interest is expressed in mammalian cells as a kinase-NanoLuc® fusion construct. As NanoLuc® Luciferase is extremely bright, only low expression of the kinase-NanoLuc® fusion protein is needed.
A cell-permeable fluorescent NanoBRET™ tracer is designed by tagging a compound of interest (typically, a known inhibitor) with a fluorescent dye. This tracer is added to the cells, followed by a NanoLuc® substrate and an extracellular NanoLuc® inhibitor. Expression of the kinase-NanoLuc® fusion allows a strong BRET signal to be achieved between the kinase-NanoLuc® protein and NanoBRET™ tracer. The extracellular NanoLuc® inhibitor ensures that the BRET signal achieved is from live, uncompromised cells. The presence of unlabeled test compound that binds to the target kinase results in a loss of BRET signal. Because BRET has tight distance constraints, the data obtained are specific for the kinase fused to NanoLuc® Luciferase.
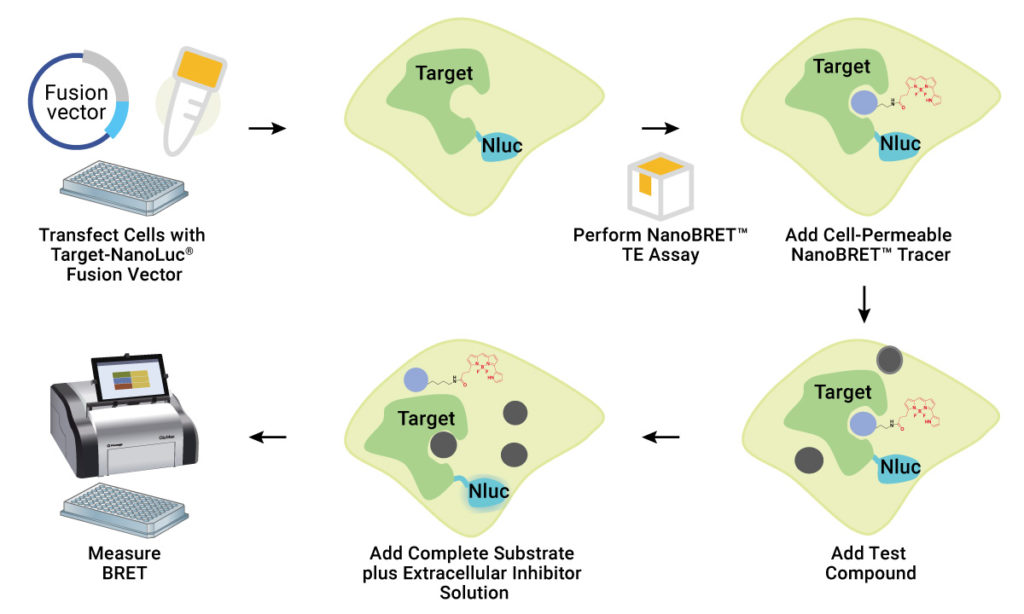
NanoBRET™ TE assays provide a sensitive, high-throughput method to study kinase target engagement.
NanoBRET™ TE assays have been used to study the interactions of a wide variety of kinases and test compounds. A previous blog post discusses the use of this technology to evaluate cyclin-dependent kinase inhibitors (9). A few other recent examples are highlighted here.
ALK4/ALK5
The transforming growth factor beta-receptor I/activin receptor-like kinase 5 (TGFBR1/ALK5), and its close homolog ALK4, are receptor protein kinases associated with the development of multiple diseases. Commercially available inhibitors either lack selectivity or have not been comprehensively characterized. Hanke et al. characterized a chemical probe, TP-008, with dual activity for ALK4 and ALK5, as well as related negative control compounds using the NanoBRET™ TE assay (10). They observed that TP-008 displayed the best selectivity of all known ALK5 inhibitors, making it an excellent tool to study the ALK4/5 signaling pathway.
SIK2/SIK3
Jin et al. studied isoforms of the salt-inducible kinases (SIKs) that modulate the production of pro- and anti-inflammatory cytokines in immune cells (11). They screened a library of 530 compounds to measure differential engagement with two SIK isoforms (SIK2 and SIK3) using NanoBRET™ TE assays in a high-throughput format and confirmed the results with time-resolved FRET assays. Their results demonstrate the sensitivity of the NanoBRET™ assay and its utility for high-throughput small-molecule TE screening.
ATP Mimetics
Since protein kinases contain a common ATP-binding site, one drug design strategy is to test ATP-mimetic inhibitors that bind to this site. However, such inhibitors display poor selectivity. Schroder et al. studied the DFG motif commonly targeted by these inhibitors and developed a rational drug design approach by including residues N-terminal to the DFG motif (12). Based on the results of NanoBRET™ TE assays and biophysical analyses, they state that the improved activities and selectivity profiles of the tested inhibitors demonstrate the validity of their approach and its utility for the design of chemical libraries targeting many kinases.
Dark Kinases
“Dark” or understudied kinases are those whose biological function is poorly understood. Tamir et al. conducted kinome-wide screening assays for 403 wild-type human kinases using the inhibitor GW296115 (13). Their results identified several dark kinases, as well as the brain-specific kinases BRSK1 and BRSK2, for which GW296115 was a potent inhibitor. A drug design strategy developed by Drewry et al., targeting the kinase pyrimidine hinge-binding motif, also led to the identification of inhibitors for several dark kinases (14). An earlier blog post describes the development of a highly selective chemical probe for the dark kinase Doublecortin-like kinase 1 (DCLK1) (15).
Compound Selectivity Profiling Services
A publication by Robers et al. describes modification of the original NanoBRET™ TE kinase profiling workflow to use a single kinase tracer, instead of six (16). The tracer can be used at four different concentrations, and the protocol demonstrates the use of this method to measure target engagement for a panel of 192 kinases with the kinase inhibitor crizotinib.
For researchers who wish to outsource their compound selectivity profiling experiments, Promega now offers a quarterly profiling service. Client-supplied compounds are tested against comprehensive panels of full-length kinases (custom kinase profiling is available on request). The profiling data include information about cellular selectivity and off-target activity.
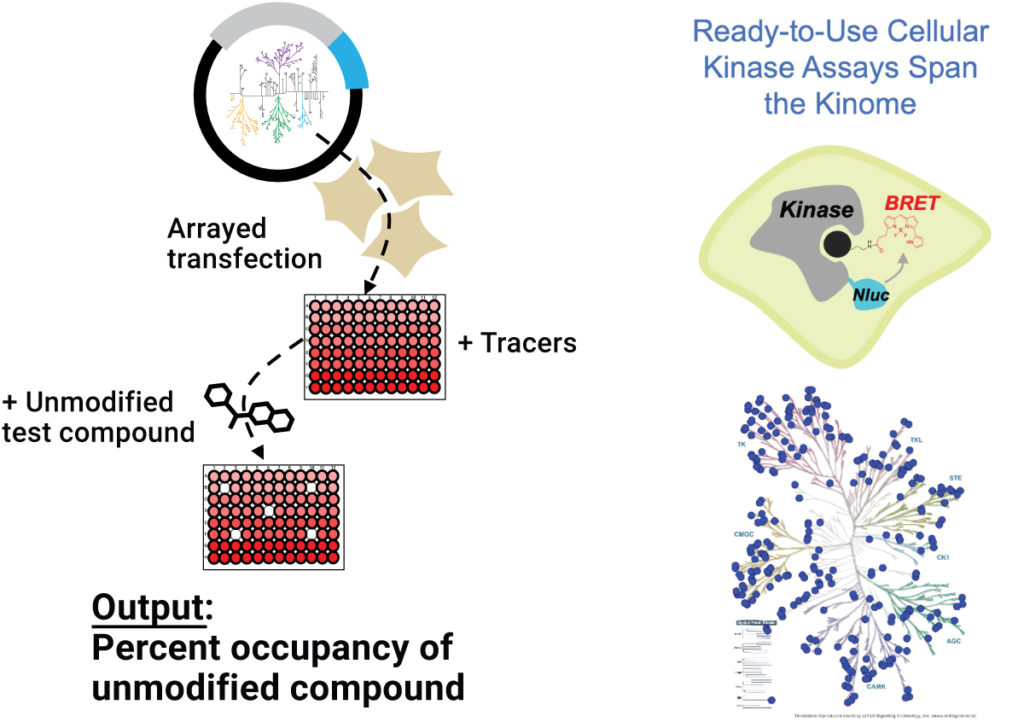
Overview of the kinase selectivity profiling service.
To learn more about profiling compound selectivity against 192 kinases in live cells, see the Selectivity System product page.
References
- Zhang, H. et al. (2021) A subcellular map of the human kinome. eLife 10:e64943 DOI: 10.7554/eLife.64943.
- Simatou, A. et al. (2020) Historical retrospective of the SRC oncogene and new perspectives (Review). Mol. Clin. Oncol. 13, 21.
- Cohen, P. et al. (2021) Kinase drug discovery 20 years after imatinib: progress and future directions. Nat. Rev. Drug Discov. 20, 551–569.
- Cho, E.J. and Dalby, K.N. (2021) SLAS Discov. Luminescence energy transfer–based screening and target engagement approaches for chemical biology and drug discovery. 26(8) 984–994.
- Xu, Y. et al. (1999) A bioluminescence resonance energy transfer (BRET) system: Application to interacting circadian clock proteins. Proc. Natl. Acad. Sci. USA 96, 151–156.
- Pfleger, K.D. and Eidne, K.A. (2006) Illuminating insights into protein-protein interactions using bioluminescence resonance energy transfer (BRET). Nat. Methods 3, 165–174.
- Hall, M.P. et al. (2012) Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol. 7, 1848−1857.
- Machleidt, T. et al. (2015) NanoBRET—A novel BRET platform for the analysis of protein-protein interactions. ACS Chem. Biol. 10, 1797−1804.
- Wells, C.I. et al. (2020) Quantifying CDK inhibitor selectivity in live cells. Nat. Comm. 11, 2743.
- Hanke, T. et al. (2020) A highly selective chemical probe for activin receptor-like kinases ALK4 and ALK5. ACS Chem. Biol. 15, 862−870.
- Jin, H.Y. et al. (2020) High-throughput implementation of the NanoBRET target engagement intracellular kinase assay to reveal differential compound engagement by SIK2/3 isoforms. SLAS Discov. 25(2) 215–222.
- Schröder, M. et al. (2020) DFG‑1 residue controls inhibitor binding mode and affinity, providing a basis for rational design of kinase inhibitor selectivity. J. Med. Chem. 63, 10224−10234.
- Tamir, T.Y. et al. (2020) PKIS deep dive yields a chemical starting point for dark kinases and a cell active BRSK2 inhibitor. Sci. Rep. 10, 15826.
- Drewry, D.H. et al. (2021) Identification of pyrimidine-based lead compounds for understudied kinases implicated in driving neurodegeneration. J. Med. Chem. (preprint) https://doi.org/10.1021/acs.jmedchem.1c00440
- Ferguson, F. M. et al. (2020) Discovery of a selective inhibitor of doublecortin like kinase 1. Nat. Chem. Biol. 16, 635–643.
- Robers, M.B. et al. (2021) Single tracer-based protocol for broad spectrum kinase profiling in live cells with NanoBRET. STAR Protocols 2(4), 100822.
Latest posts by Ken Doyle (see all)
- Will Artificial Intelligence (AI) Transform the Future of Life Science Research? - February 1, 2024
- RAF Inhibitors: Quantifying Drug-Target Occupancy at Active RAS-RAF Complexes in Live Cells - September 5, 2023
- Synthetic Biology: Minimal Cell, Maximal Opportunity - July 25, 2023
