Part two of three. You can read part 1 here.
Formalin-Fixed Paraffin embedded (FFPE) samples are being used in increasing numbers of molecular assays. In my last blog I discussed some of the pre-analytical variables that can affect results obtained when using FFPE samples. Laboratories can increase the quality of downstream results by controlling variables where possible. While exacting control over the sample acquisition and fixation process can improve results, quality testing of incoming samples is a crucial step in assuring optimal results. There are numerous methods that can be used to evaluate the quality of samples and they can provide different information that can be used to assess sample integrity and suitability for different applications.
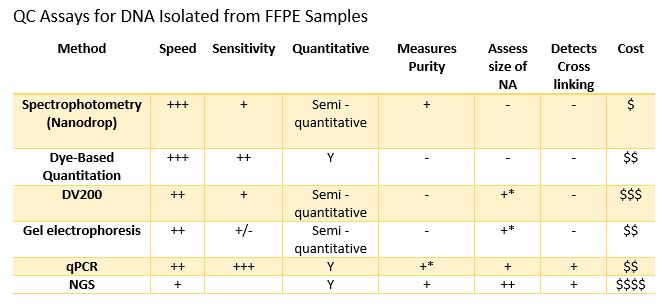
Spectrophotometry is a common way to evaluate the quality of extracted DNA and RNA. Most laboratories have a NanoDrop Microvolume Spectrophotometer (or similar device), and they are incredibly easy to use. Pipette 1-2µl of sample, select “Analyze” the instrument provides a readout of concentration and purity via A260/A280 and A260/A230 ratios in a few seconds. These devices have revolutionized routine sample quantitation in the lab, but is it the best method for assessing FFPE samples? There are two considerations when using Nanodrop analysis: sensitivity and integrity. FFPE samples can have a wide-ranging yield of DNA or RNA, often as little as 10ng or less in a volume ranging from 10µl to 100µl from an extraction. This can result in sample concentrations below the linear range of the instrument. In addition, spectrophotometers ike the Nanodrop do not differentiate between RNA, DNA or free nucleotides, which can result in dramatic inaccuracies in DNA / RNA concentration measurements. Finally, there is no way to determine if a sample is accessible to downstream enzymatic assays since it cannot detect the presence or absence of crosslinks (or other damage) within a sample.
Dye based Quantitation like the Promega QuantiFluor® dsDNA System, provides a rapid and significantly more sensitive method to quantitate dsDNA or RNA compared to absorbance spectroscopy. This method provides a broadly useful estimate of concentration. When considering FFPE samples, it is important to note that dye-based quantitation does not estimate the integrity of the DNA/RNA or the extent of cross linking in the sample, which could affect success in downstream assays.
Sizing assays (e.g., agarose gel, Tape station, fragment analyzer, DV200) can provide an estimate of concentration and more importantly information on the size distribution of the fragments in the sample. FFPE-derived DNA, due to the fixation process, can be significantly fragmented compared to DNA from freshly frozen tissue. Below is a fragment analyzer trace and associated DV200 scores of DNA isolated from FFPE sections using five different purification methods. While the sizing traces do assess the distribution of DNA size purified, it does not measure the degree of cross linking within the sample or the presence of inhibitors.
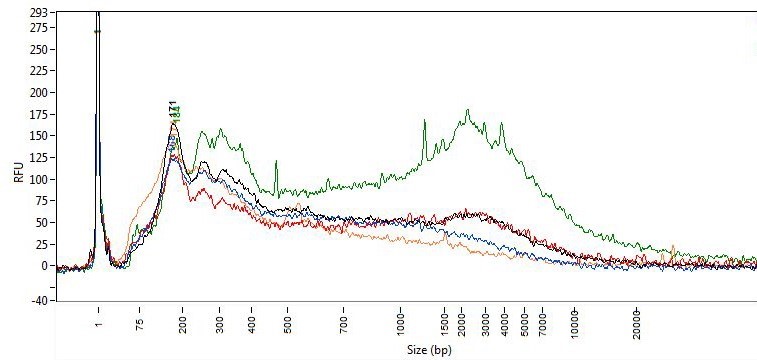
DV200 Scores
| Method | 1 | 2 | 3 | 4 | 5 |
| DV200 | 71.8 | 69.9 | 70.1 | 58.7 | 80.5 |
When the same samples were quantitated by qPCR assays of various targets and fragment sizes, the yield by qPCR does not correlate well with the DV 200 scores. In fact, in this example, the samples with the lowest DV200 scores had the greatest yield by qPCR (data below ). While there are general trends, the DV200 score does not necessarily correlate with success in downstream assays such as qPCR.
qPCR has several advantages for the quantitation of FFPE samples. First, qPCR can be very sensitive requiring only a small amount of sample and, detecting pg/µl amounts of DNA. In terms of sensitivity in detecting nucleic acids, it is only surpassed by ddPCR. qPCR also can provide a measure of how degraded or crosslinked a DNA sample may be since nucleic acid must be a suitable substrate for a DNA polymerase for a signal to be generated. Absorbance may not represent the sample suitable for the downstream assay because it will detect DNA fragmented DNA and nucleotides. Finally, most qPCR QC assays such as the ProNex® DNA QC Assay provide internal controls which are used to detect the presence of inhibitors in the sample prior to attempting a more expensive assay. This can help you assess not only the integrity of the nucleic acids but also the likelihood of an amplification-based assay to be successful.
NGS is used by some labs to QC their samples. There are several reasons for this. Some labs are trying to get as much data as possible from very precious samples, in which case any sequence information may be worth the expense and risk of failed sequencing runs. As a QC test, NGS may provide lots of information, but it is expensive and can require large amounts of sample and time. Most sequencing and purification providers recommend qPCR assays to quantitate FFPE DNA. Others run low pass NGS, which uses highly multiplexed samples to lower the cost per sample to determine if it is worth the time and sequencing resources to sequence deeper. Most sequencing and purification providers recommend qPCR assays to quantitate FFPE DNA because all NGS workflows depend primarily on the success of enzymatic amplification steps to obtain sequencing-ready DNA as part of library prep steps.
To learn more about how a qPCR assay can predict NGS success, view my colleague Doug Wieczorek’s poster, Purification, Quantitation and QC Measurements for Predicting Downstream NGS Success with FFPE and Circulating Cell-Free DNA Plasma Samples.
Another challenge faced by laboratories is sample throughput. As molecular assays become more informative or are being considered for companion diagnostic assays, the number of FFPE samples being processed for DNA or RNA analysis is increasing dramatically. In my final blog, I’ll discuss some of the challenges with automating FFPE sample processing and ways that Promega has automated FFPE extractions on the Maxwell RSC and on third-party liquid handlers.
