Studying protein function in live cells is limited by the tools available to analyze the expression and interactions of those proteins. Although mass spectrometry and antibody-based protein detection are valuable technologies for protein analysis, both methods have drawbacks that limit the range of targets and contexts in which proteins can be investigated.
Mass spectrometry is often poor at detecting low-abundance proteins. Antibody-based techniques require high quality, specific antibodies, which can be difficult to impossible to acquire. Both methods require cell lysis, preventing real-time analysis and limiting the physiological relevance, and both methods can be limiting for higher-throughput analysis. While plasmid-based overexpression of tagged target proteins simplifies detection and can allow for real time analysis, protein levels don’t typically resemble endogenous levels. Overexpression also has the potential to create experimental artifacts or limit the dynamic range of an observed response.
In 2018, Promega R&D scientists published a paper in ACS Chemical Biology demonstrating the use of CRISPR/Cas9 to integrate the 11 amino acid, bioluminescent HiBiT tag directly into the genome to serve as an easily measured reporter for endogenous proteins. This provides a highly quantitative method for investigating cellular protein dynamics that sidesteps the need for cloning and other drawbacks to conventional methods, including the ability to measure changing protein dynamics in real-time. (For more details about CRISPR/Cas9 knock-in tagging and other applications, read this blog.)
While their findings showed that this method provides efficient and specific tagging of endogenous proteins, the research was limited to just five different proteins within a single signaling pathway in two cell lines. This left unanswered questions about whether this approach was scalable, had broader applications and how accurately the natural biology of the cells was represented.
The latest issue of Scientific Reports features new research from Promega R&D scientists that builds on the original paper by doing CRISPR/Cas9 knock-in of the HiBiT reporter tag on a more comprehensive and diverse set of proteins in a few different cells lines. The researchers selected 97 targets (in four cell lines) that were diverse in size, subcellular localization, physiological function and transcript expression levels. (You can find the guide RNA and donor DNA sequences for all of the proteins that were tagged in the study in this supplement.)
The researchers showed that 83 of the targets could be HiBiT tagged and detected by luminescence. Using Western blotting to investigate these targets would require 97 antibodies, making it more expensive and time-consuming; furthermore, it may not even be possible to conduct the experiment if an antibody doesn’t exist for a particular target. Their findings demonstrate that this approach is a broadly applicable and scalable method to analyze endogenous proteins.
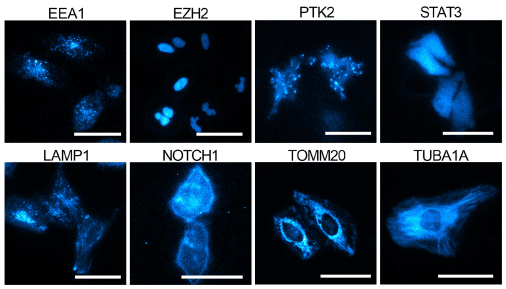
This report also provides greater detail on the validation and characterization of the knock-in cells. After screening edited cell pools for luminescence, additional validation to confirm expression, size, and localization of the HiBiT tagged proteins was performed. HiBiT-specific blotting and bioluminescence imaging techniques were used in lieu of Western blotting and immunofluorescence microscopy. HiBiT’s sensitivity allows multi-format, quantitative analysis in luminescence plate assays, blotting, and bioluminescence imaging.
Finally, the report aims to better understand the performance of cell models that are developed with endogenous protein tagging. By comparing data for HiBiT fusions expressed from the endogenous loci and from a plasmid of two transcriptionally regulated proteins, the researchers demonstrated the advantage of using edited cells over traditional overexpression models. Only endogenous expression provided data that accurately captured cellular protein dynamics. Preserving cell integrity allows signaling pathways to be analyzed in a natural state, free of the artifacts associated with overexpression, and edited cells better represent target biology.
Want to see how we have used this technique to build a catalog of available cell lines or get information on how to do it yourself? Learn more
Related Posts
Latest posts by Darcia Schweitzer (see all)
- Cytochrome P450 Inhibition: Old Drug, New Tricks - May 5, 2022
- Firefly Luciferase Sheds Light on Development of New Malaria Treatments - April 5, 2021
- How to Train Your Instrument Service Team in a Pandemic - February 1, 2021
