I was having a discussion with my mother just the other day about cleaning products (lively topic, I know). She showed me her newest time saver…prediluted bleach. Huh, I thought. I guess that does save a bit of time, but I couldn’t resist telling her that she was paying triple the price for a whole lot of water. She said, without pause, that it was worth it to her to not have to splash fully concentrated bleach around. A convenience worth paying for, in her words.
I don’t know why this struck me as odd. I pay for convenience all the time as I get older. When I started running gels back in college, I wouldn’t have dreamed of buying a precast gel, but several years into my lab life I found myself running more than 15 gels a week, so precast was really a convenient alternative. When I was a grad student, I poured all of my own plates (and most of the plates for older students, too!). Fast forward a few years, and I running upwards of 300 microbial selective cultures per week. The switch to prepoured plates was a no brainer.
When put in the context of what our time is worth, would you rather be thawing and mixing loading dyes, buffers, stains, reagents, etc., or are you better of grabbing a premixed, room-temp stable dye or ladder/loading dye mix off the shelf and getting on with your research? I think most scientists would agree that these small conveniences allow you to free up a little more time to do the important work you should be doing.
I’m curious…what time savers or convenience items do you find that make your day a little easier in the lab?
Like this:
Like Loading...




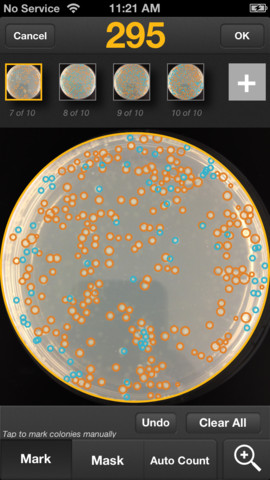 Do you count colonies on agar plates? Do you often need to average counts over a series of plates? The
Do you count colonies on agar plates? Do you often need to average counts over a series of plates? The