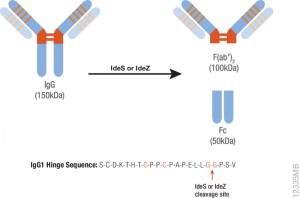
During preclinical research and development of therapeutic antibodies, multiple variants of each antibody are assessed for pharmacokinetic (PK) characteristics across model systems such as rodents, beagles and primates. Ligand-binding assays (LBA) or liquid chromatography coupled to tandem mass spectrometry(LC–MS/MS)-based methods represent the two most common technologies used to perform the PK studies for mAb candidates(1,2).
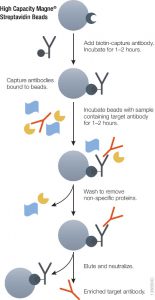
Using either method it is essential to ensure accurate quantitative results that the initial enrichment of the target therapeutic antibody from serum or plasma be optimal. Biotinylated antibodies or antigens (against the therapeutic targets) immobilized onto high capacity streptavidin beads will enrich therapeutic antibody from serum or plasma samples. The affinity of biotin for streptavidin (Kd = 10–15) is one of the strongest and most stable interactions in biology therefore the biotin-streptavidin interaction cannot be reversed under non-denaturing conditions. Hence, it is possible to perform extensive washing to remove nonspecifically bound protein and elute therapeutic antibodies without also eluting the biotinylated component, thus improving the detection limit.
Magnetic based separation techniques have several advantages in comparison with standard separation procedures. This process is usually very simple, with only a few handling steps. All the steps of the purification procedure can take place in one single test tube. The magnetic separation techniques are also the basis of various automated procedures. Learn more about the High Capacity Magne™ Streptavidin Beads (Cat # V7820) .
References
- DeSilva, B. et al. (2003) Recommendations for the Bioanalytical Method Validation of Ligand-Binding Assays to Support Pharmacokinetic Assessments of Macromolecules. Pharm. Res. 20, 1885–00.
- Zhang, Q. et al. (2014) Generic Automated Method for Liquid Chromatography–Multiple Reaction Monitoring Mass Spectrometry Based Monoclonal Antibody Quantitation for Preclinical Pharmacokinetic Studies. Anal. Chem. 86, 8776–84.
 Cerebrospinal fluid (CSF) is a bodily fluid present around the brain and in the spinal cord. It acts as a protective cushion against shocks and participates in the immune response in the brain. Analysis of total CSF protein can be used for diagnostic purposes, as, for instance, a sign of a tumor, bleeding, inflammation, or injury. Considering the high value of CSF as a source of potential biomarkers for brain-associated damages and pathologies, the development of robust automated platform for CSF proteomics is of great value.
Cerebrospinal fluid (CSF) is a bodily fluid present around the brain and in the spinal cord. It acts as a protective cushion against shocks and participates in the immune response in the brain. Analysis of total CSF protein can be used for diagnostic purposes, as, for instance, a sign of a tumor, bleeding, inflammation, or injury. Considering the high value of CSF as a source of potential biomarkers for brain-associated damages and pathologies, the development of robust automated platform for CSF proteomics is of great value.