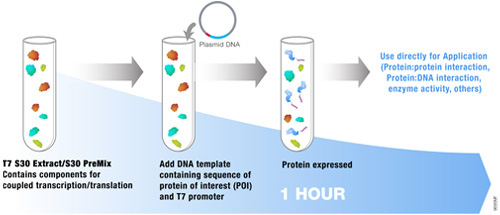
Cell-free systems can provide some advantages over cell-based systems for screening purposes. Cell-free systems allow exact manipulation of compound concentrations. This is an important parameter when evaluating the potential potency of the lead compound.
There is no need for cellular uptake to evaluate the effect of the compounds. While uptake evaluation is important for determining the eventual efficacy of the drug, it can unnecessarily eliminate valuable lead compounds in an initial screen. The interpretation of results in living cells is complicated by the large number of intertwined biochemical pathways and the ever-changing landscape of the growing cell. Cell-free systems allow the dissection of effects in a static system for simpler interpretation of results and the ability to specifically monitor individual processes such as transcription or translation. Individual targets not normally present, or found at low concentrations, can be added in controlled amounts.
The following references illustrate this application:
- Buurman, E. T. et al. (2012) Novel rapidly diversifiable antimicrobial RNA polymerase switch region inhibitors with confirmed mode of action in Haemophilus influenzae. J. Bacteriology 194, 5504–12.
-
Zhang, Y. and Inouye, M (2009) The inhibitory mechanism of protein synthesis by YoeB, an Escherichia coli toxin. J. Biol. Chem. 284, 6627–38.
- Jayasekera, M.M. et al. (2005) Identification of novel inhibitors of bacterial translation elongation factors Antimicrob. Agents Chemother. 49, 131-36.