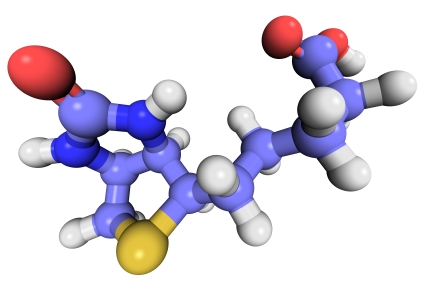
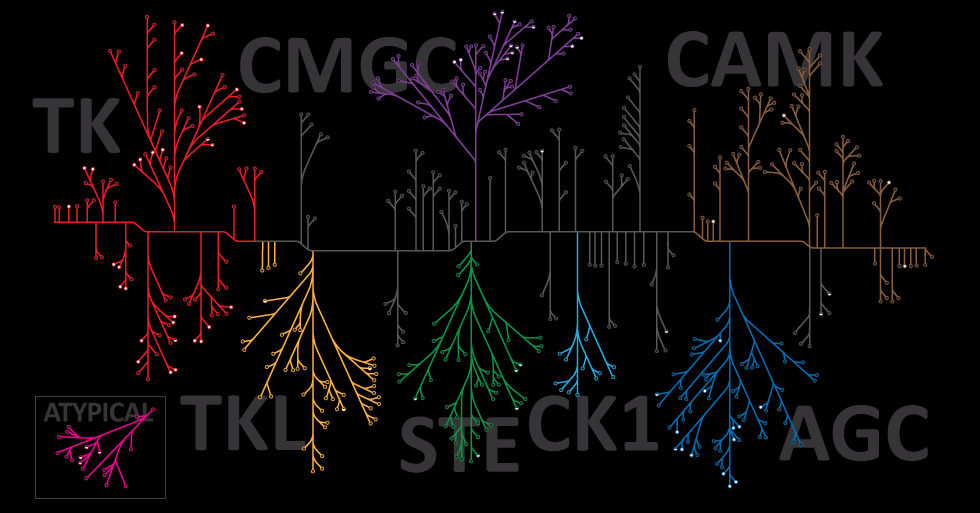
 Protein phosphorylation is one of the most biologically relevant modifications and is involved in many eukaryotic and prokaryotic cellular signaling processes. It is estimated that one-third of human proteins are phosphorylated.
Protein phosphorylation is one of the most biologically relevant modifications and is involved in many eukaryotic and prokaryotic cellular signaling processes. It is estimated that one-third of human proteins are phosphorylated.
The following examples utilize the ability of cell free experession to express active proteins, and when supplemented with the necessary components (e.g., ATP, NaCl), to be used for the characterization of phosphorylation events.
Modrof, J. et al. (2005) Phosphorylation of bluetongue virus nonstructural protein 2 is essential for formation of viral inclusion bodies. J. Vir. 79, 10023–31. Use of TNT® cell-free to express NS2 and NS2 mutant proteins for use in vitro kinase assays to confirm phosphorylation by protein kinase CK2.
Kwon, S. et al. (2005) Signal pathway of hypoxia-inducible factor-1alpha phosphorylation and its interaction with von Hippel-Lindau tumor suppressor protein during ischemia in MiaPaCa-2 pancreatic cancer cells. Clin. Cancer Res. 11, 7607–13. The TNT® system was used to identify which p38 mitogen-activated protein kinase isoform(s) was cabable of phosphorylation of HIF—1 alpha
Harris, J. et al. (2006). Nuclear accumulation of cRel following C-terminal phosphorylation by TBK1/IKK epsilon. J. Immunol. 177, 2527–35. IKK and IKK mutants were expressed using TNT and used in a vitro kinase assay to characterize the recognition motif in cRel transcription domain
Jailais, Y. et al. (2011) Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev. 25, 232–37. Using a vitro kinase assay, full–length and truncations versions of the Brassinostediod-insentive receptor protein were expressed using the TNT® system and incubated with purified BR11 kinase domain to determine binding sites of the two proteins.
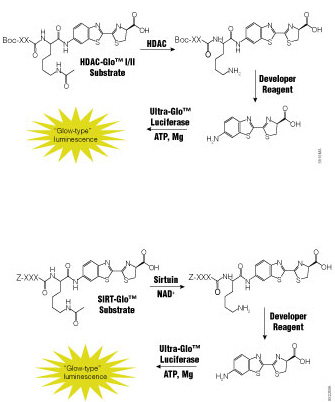


 The ability to manipulate genes and proteins and observe the effects of specific changes is a foundational aspect of molecular biology. From the first site-directed mutagenesis systems to the development of knockout mice and RNA interference, technologies for making targeted changes to specific proteins to eliminate their expression or alter their function have made tremendous contributions to scientific discovery.
The ability to manipulate genes and proteins and observe the effects of specific changes is a foundational aspect of molecular biology. From the first site-directed mutagenesis systems to the development of knockout mice and RNA interference, technologies for making targeted changes to specific proteins to eliminate their expression or alter their function have made tremendous contributions to scientific discovery.