Today’s blog post is written by guest blogger Sarah Kolb, Marketing Coordinator for our North America Branch, and new employee at Promega.
As a new member to the North America Marketing team, I was unsure of what to expect going into my first national sales meeting with Promega, but what I took away from this meeting was incredibly eye opening. The North America Branch Sales meeting is an opportunity to get all of the members of the North American branch together to learn about new products, connect with the different strategic business units about product application and network with each other to learn how to better the lives of our customers. The year’s meeting occurred in May in the Ideation room at Promega Headquarters in Madison, Wisconsin.
The room itself is not your typical conference space. An antique car resides in the space, and you can find art-work from all over world nestled in corners, and on the walls and shelves. All around the room, collections of unique furniture are arranged to stimulate conversation. Ideation created an atmosphere of creativity, community and collaboration, which contributed to the overall success of the meeting.
I felt excitement Monday morning as the 62 attendees gathered in the space. Everyone greeted each other with big smiles and hugs, asking about families, travels and already discussing business. Continue reading “What I Learned at My First Branch Meeting: Gratitude, Service and Collaboration”

 The product’s launch was an exciting milestone for Promega as research interest in the role of ccfDNA as biomarkers in human disease continues to grow. Elevated levels of ccfDNA have now been reported in patients with cancer, inflammatory disease, infections and cardiovascular disease. In pregnant women, up to 10% of ccfDNA can be attributed to the fetus, so critical fetal DNA analysis can now be conducted through maternal blood samples. There are many advantages in the ability to isolate and analyze ccfDNA, so the development of a kit with high throughput capability was a priority for the Nucleic Acid Purification R&D team.
The product’s launch was an exciting milestone for Promega as research interest in the role of ccfDNA as biomarkers in human disease continues to grow. Elevated levels of ccfDNA have now been reported in patients with cancer, inflammatory disease, infections and cardiovascular disease. In pregnant women, up to 10% of ccfDNA can be attributed to the fetus, so critical fetal DNA analysis can now be conducted through maternal blood samples. There are many advantages in the ability to isolate and analyze ccfDNA, so the development of a kit with high throughput capability was a priority for the Nucleic Acid Purification R&D team. 
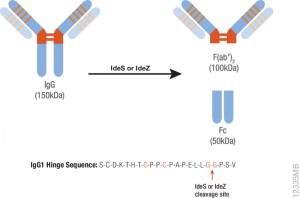
 Antibodies labeled with small molecules such as fluorophore, biotin or drugs play a critical role in various areas of biological research,drug discovery and diagnostics. There are several limitations to current methods for labeling antibodies including the need for purified antibodies at high concentrations and multiple buffer exchange steps.
Antibodies labeled with small molecules such as fluorophore, biotin or drugs play a critical role in various areas of biological research,drug discovery and diagnostics. There are several limitations to current methods for labeling antibodies including the need for purified antibodies at high concentrations and multiple buffer exchange steps. Ricin, derived from caster seeds, inhibits protein synthesis by binding to ribosomes, resulting in cell death. The protein is composed of two polypeptide chains: Ricin Toxin A chain and Ricin Toxin B chain. Ricin inhibits protein synthesis very quickly, and the cell or tissue damage begins within several hours. However, signs of poisoning often are not noted before significant damage has been done, making treatment difficult. Therapeutics that either block the ribosome binding site or compete with the toxin for binding are highly desired. Both antibodies and competitive ligands inhibited binding of the toxin to cell membranes.
Ricin, derived from caster seeds, inhibits protein synthesis by binding to ribosomes, resulting in cell death. The protein is composed of two polypeptide chains: Ricin Toxin A chain and Ricin Toxin B chain. Ricin inhibits protein synthesis very quickly, and the cell or tissue damage begins within several hours. However, signs of poisoning often are not noted before significant damage has been done, making treatment difficult. Therapeutics that either block the ribosome binding site or compete with the toxin for binding are highly desired. Both antibodies and competitive ligands inhibited binding of the toxin to cell membranes.