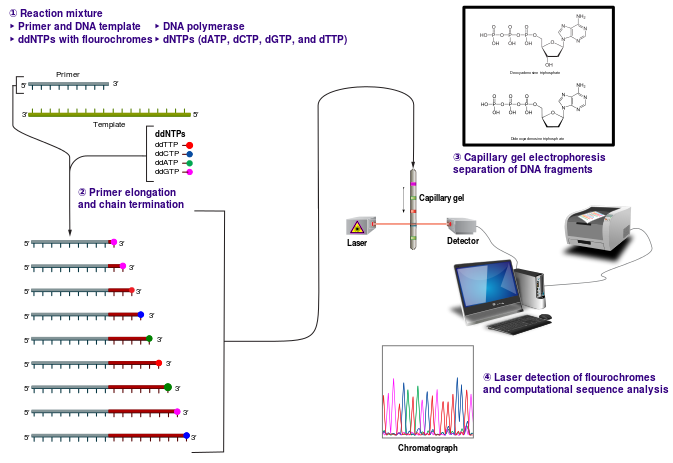
We’ve learned this year, 2013, that Europe may be the original home to domestic dogs, a title previously claimed by East Asia and the Middle East. A recent study published in Science magazine may put to rest the debate.
In their report, Thalmann and Wayne (1) used an evidential gold standard, DNA from mitochondria of fossilized ancient dog and wolf remains, to reach the conclusion that dogs originated from a now extinct line of European gray wolves.
In 2002, researchers from Sweden and China collaborated to compared first the mitochondrial DNA and later the complete mitochondrial genomes and Y chromosomes from a hundreds of wolves, coyotes and modern dogs from around the world (2). Their results showed the greatest genetic diversity from canids from East Asia. Such genetic diversity can be a marker of a species’ origin.
In 2010 Wayne, et al. (3) analyzed 48,000 markers from across the genome of gray wolves and dogs, again from around the world. The dogs were found to have more genetic material in common with Middle Eastern wolves than with those from East Asia. Wayne and colleagues found this a yes to dogs’ origins lying in the Middle East.
A criticism of the analysis done with modern dog DNA is that this DNA has been mixed with that from wolves. In addition, dog-dog breeding over the 15,000 to 30,000 years since this domestication, could confound results. Prior to the current study, critics called for analysis of ancient DNA remains only.
Wayne had collected DNA from ancient remains and due to recent collaboration with geneticist Thalmann, now had the lab-power to analyze those remains.
In this work, Wayne and Thalmann looked at mitochondrial DNA from ancient remains, those of 18 wolves and dogs. The fossils ranging in age from 1,000 to 36,000 years. Eight of the samples were classified as dog-like and 10 samples were wolf-like in nature. They compared the ancient mitochondrial DNA samples to those of modern animals, including 77 dogs from an assortment of breeds, as well as 49 wolves and 4 coyotes. They then built a sort of a canid family tree, demonstrating relatedness in the animals whose DNA was analyzed.
Thalmann and Wayne’s finding showed that 1) the DNA of modern dogs more closely resembled that of ancient gray wolves than modern wolves, and 2) the geographic location of the wolves who’s DNA was most closely resembled, was Europe.
It is important to note that Thalmann and Wayne did not compare ancient remains from animals from the Middle East, nor did they have access to ancient remains from East Asia. In addition, there is criticism of the use of mitochondrial DNA, which represents only the maternal dog lineage. Thus there is more work to be done to finalize the ancestral home of the modern dog.
This work does, however, push back the origins of domesticated dogs to between 18,000 and 32,000 years ago, significant because the domestication timeline was previously believed to coincide with the rise of farming by our human ancestors. Domestic canines are now believed to have been part of humans lives far before farming was a way of life, back in the hunter-gatherer days.
There are those who believe that domestication of wolves, selectively bred to become modern dogs occurred simultaneously at more than one geographic region. Wolves were once found in many locations around the world and their usefulness to and domestication by humans would seem odd if only embraced by people from a single part of the world.
References
- Thalmann, O. et al. Science (2013) 342, 871–874
- Savolainen P, Zhang YP, et al. (2002)
- Wayne, R.K. et al. (2010) Nature 464(7290), 898-902.
Like this:
Like Loading...



 Paul Steyn has posted an
Paul Steyn has posted an