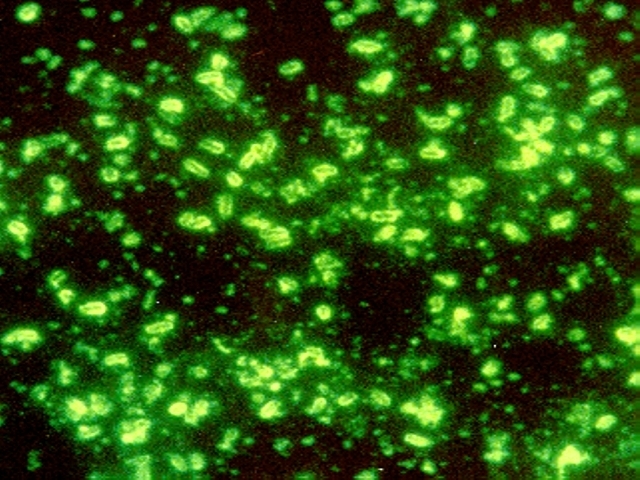
When I started writing about research on Yersinia pestis and the Black Death, I was amazed at the ability to recover 14th century bacterial DNA from human remains, show Y. pestis was the caustive agent of the Black Death and then sequence the strain to compare to modern Y. pestis strains. The publications I read always mentioned the three waves of pandemics that devastated human populations in the introduction, and the Black Death was not the oldest one. The putative first pandemic was the Plague of Justinian in the 6th century, named after the Byzantine emperor. Like with the Black Death, there is debate about whether Y. pestis is the causative agent of the Plague of Justinian. The research published in PLOS Pathogens built on earlier work to isolate and genotype the suspected Y. pestis causative agent from human remains in 6th century graves, but this time with more stringent protocols enacted to answer critics who questioned the authenticity of earlier results.
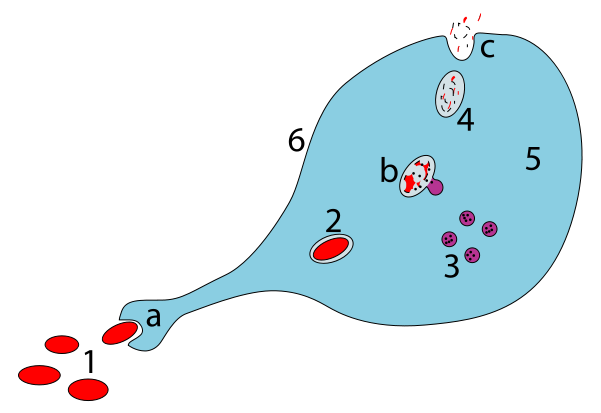



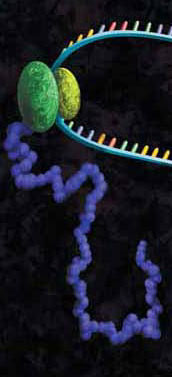
 A little over a year ago, I wrote about
A little over a year ago, I wrote about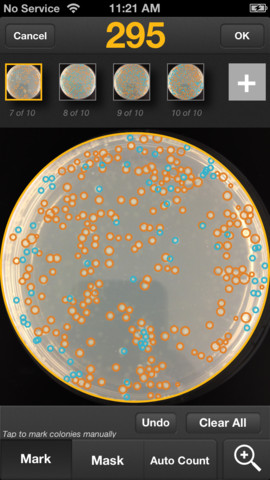 Do you count colonies on agar plates? Do you often need to average counts over a series of plates? The
Do you count colonies on agar plates? Do you often need to average counts over a series of plates? The 
 Stainless steel is often used in clinical and public settings as work surfaces as well as other surfaces that are touched and cleaned often. Stainless steel is used in these applications for many of the same reasons I like it for jewelry: it is strong, resilient, relatively inexpensive, stain- and corrosion-resistant and will weather regular cleaning/exposure to moisture well. There is something about a gleaming stainless steel work surface that looks, well, sterile. But is it?
Stainless steel is often used in clinical and public settings as work surfaces as well as other surfaces that are touched and cleaned often. Stainless steel is used in these applications for many of the same reasons I like it for jewelry: it is strong, resilient, relatively inexpensive, stain- and corrosion-resistant and will weather regular cleaning/exposure to moisture well. There is something about a gleaming stainless steel work surface that looks, well, sterile. But is it? 