Microorganisms: Infectious Disease and Ecology
Blog entries about bacteria, viruses, fungi and other microorganisms.
Congenital Cytomegalovirus: The Most Common Congenital Infection That You Have Never Heard Of

Down Syndrome. Fetal alcohol syndrome. Spina bifida. Most people have heard of these well know congenital conditions, and know at least in a general sense that they have profound and lasting effects on the children born with them as well as their families. Unfortunately, people are much less aware of a congenital infection that is more common that any of these and affects more infants than all three of the conditions listed above. In fact, this congenital infection causes more cases of congenital disease than all of the 29 conditions currently screened for in most American states combined (1; 2), and yet it is not widely known by the general public. Cytomegalovirus, or CMV, is the most common congenital viral infection in developed countries (3; 4), and the leading cause of congenital sensorineural hearing loss and psychomotor retardation (1).
Continue reading “Congenital Cytomegalovirus: The Most Common Congenital Infection That You Have Never Heard Of”Zika Virus: Another RNA Virus Emerges

Zika virus has been in the news recently due to growing concerns about its global spread. If you have never heard of Zika virus before, you are not alone. Although first discovered in the 1940s, Zika has not been the subject of much study as infection is considered rare and the symptoms mild. However, all this has changed in recent months due to the rapid spread of the virus in Latin America, where it has been associated with an increased incidence of microcephaly, a severe birth defect where babies are born with underdeveloped brains. Although the connection of Zika with microcephaly is not yet proven, the circumstantial evidence is strong, leading the World Health Organization to declare the spread of Zika virus an international public health emergency earlier this week.
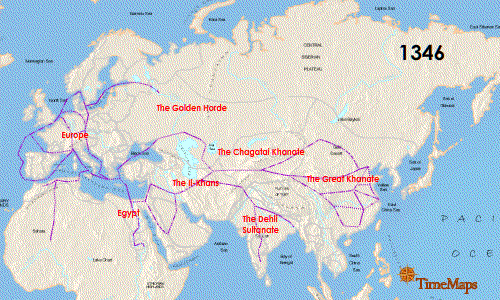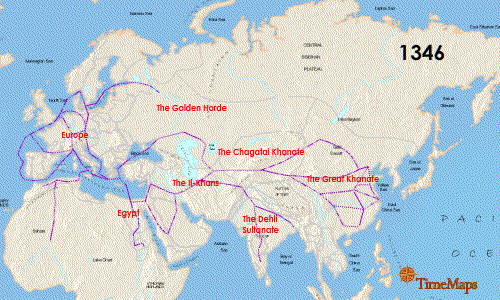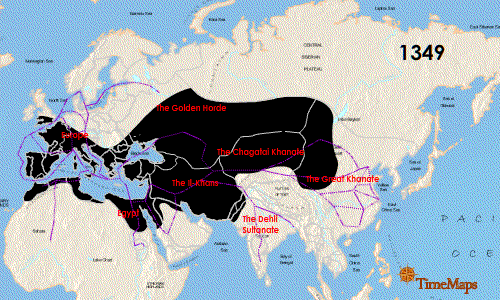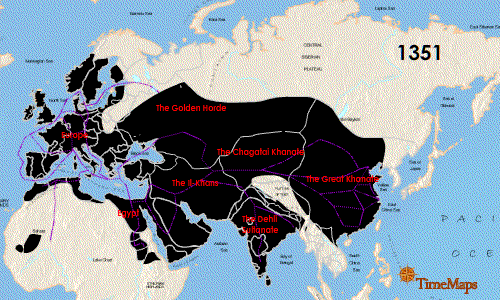
Continue reading “Zika Virus: Another RNA Virus Emerges”The Black Death: World Traveler or Persistent Homebody?
Unexpected connections: Gut bacteria influence immunotherapy outcomes
Over the last few years, human microbiome studies have revealed fascinating connections between our colonizing microorganisms and ourselves—including associations between gut bacterial populations and obesity, disease susceptibility, and even mood. The relationship between us and our microbial colonists—once considered completely benign, is now being revealed as an intricate, complicated partnership with the potential to redefine who “we” are in fundamental ways.
Two papers published back-to-back in the November 27 issue of Science add further to this growing body of knowledge—reporting a new and unexpected connection between gut bacterial species and the effectiveness of cancer immunotherapies in mice. The work suggests one reason why such treatments are effective in some circumstances, but not others. Both papers report that the presence of specific bacterial populations may be required for the efficacy of certain treatments, and raise the intriguing question “Could the composition of bacteria in the gut be manipulated to enhance the effectiveness of cancer treatments?”
Continue reading “Unexpected connections: Gut bacteria influence immunotherapy outcomes”Circulating Biomarkers and Their Applications in Gastric Cancer
 Cancer. It has been the nemesis of medical science for decades. We declare war on it, we wax philosophical about finding a cure for it. We talk about it as if it were a single enemy, but it isn’t—Cancer is not a disease, it is hundreds of diseases. These diseases manifest in every region of the human body. Many cancers, if diagnosed early, can be treated successfully. Unfortunately, there are many forms of cancers that have no external signs and very few symptoms at the early stages. Such is the enemy we face: we know what it is, we know we need to kill it early and in many cases we know how to do it, what we don’t know is how to catch it early.
Cancer. It has been the nemesis of medical science for decades. We declare war on it, we wax philosophical about finding a cure for it. We talk about it as if it were a single enemy, but it isn’t—Cancer is not a disease, it is hundreds of diseases. These diseases manifest in every region of the human body. Many cancers, if diagnosed early, can be treated successfully. Unfortunately, there are many forms of cancers that have no external signs and very few symptoms at the early stages. Such is the enemy we face: we know what it is, we know we need to kill it early and in many cases we know how to do it, what we don’t know is how to catch it early.
Gastric cancer kills approximately 745,000 people a year worldwide, making it the third most common cause of cancer-related deaths (1). It has such a high mortality rate because usually it is not detected until the disease has progressed to the later stages (IIIA–IV; 2). When detected this late, the 5-year survival rate ranges from 7–27%, with the median survival being less than 12 months (2). In contrast, when diagnosed early (i.e., cancer that is limited to the submucosal layer) it is curable with an endoscopic mucosal dissection or a minimally invasive surgery. The difference between these two outcomes is time. The earlier the cancer is detected, the better the prognosis.
Currently, upper endoscopy is the primary screening technique for detecting precancerous lesions as well as gastric cancer in the early stages. This technique has a number of downsides: it is invasive, it can have serious side effects (although these are uncommon), and it is expensive and highly dependent on the skill of the endoscopist. For these reasons, endoscopy screening is likely to suffer from poor participation rates. In addition, endoscopy is not a practical approach in low-income countries. There is clearly a need for a less invasive, sensitive screening test that will detect gastric cancer at an early stage. Continue reading “Circulating Biomarkers and Their Applications in Gastric Cancer”
Will Warmer Weather Wake the Sleeping Giant (Viruses)?
Following the discovery of Mimivirus (1) the first virus with a particles large enough to be visible under the light microscope, two additional “giant” viruses infecting Acanthamoeba have been discovered Pandoravirus (2) and Pithovirus sibericum (3), the latter from a 30,000 year old Siberian permafrost. A fourth type was recently isolated from the same sample of permafrost by Legendre et al, and named Mollivirus sibericum (4).
Mollivirus sibericum has an approximately spherical virion (0.6 µm diameter) with a 651kb GC-rich genome that encodes 523 proteins. To further characterize the virus the researchers performed transcromic- and proteomic-based time course experiments.
For the particle proteome and infectious cycle analysis, proteins were extracted and then run a 4–12% polyacrylamide gel, and trypsin digests were performed in-gel before nano LC-MS/MS analysis of the resulting peptides. Proteomic studies of the particle showed that it lacked an embarked transcription apparatus, but revealed an unusual presence of many ribosomal and ribosome-related proteins.
When the researchers explored the proteome during the course of an entire infectious cycle, the relative proportions of Mollivirus-, mitochondrion-, and Acanthamoeba encoded proteins were found to vary consistently with an infectious pattern that preserved the cellular host integrity as long as possible and with the release of newly formed virus particles through exocytosis.
In an interesting footnote, the authors of this study point out the fact that two different viruses retain their infectivity in prehistorical permafrost layers should be a concern in the context of global warming and the potential to expose humans to primeval viruses.
References
1. La Scola, B. et al. (2003) A giant virus in amoebae. Science 299, 2033.
2. Philippe, N. et al. (2013) Pandoraviruses. Amoeba virus with genomes up to 2.5Mb reaching that of parasitic eukaryotes. Science 341,281–6.
3. Legendre, M. et al. (2014) Thirty thousand year old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc.Natl. Acad. Sci. 111, 4274–9.
4. Legendre, M. et al. (2015) In depth study of Mollivirus sibercum, a new 30,000 year old giant virus infecting Acanthamoeba. Proc. Natl. Acad. Sci. 112, E5327–35 (online).
Yersinia pestis Reveals More Secrets From the Grave
Fridays are generally reserved for fun posts to share prior to the weekend. As we all know, fun is relative and to me, the latest news about how long Yersinia pestis has been entwined with human history is intriguing. I enjoy writing about the latest historical finding of Y. pestis even if I do earn a black reputation among my blogging colleagues (pun intended). Therefore, as soon as I saw the Cell article about Y. pestis found in Bronze age human teeth, I knew my blog topic was at hand.
Y. pestis has long been suspected in several plagues that occurred in the last two millennia. Publications in 2011 and 2013 used DNA extracted from teeth of human remains dated to the 14th century Black Death and 6th century Plague of Justinian to confirm Y. pestis was the causative agent in those devastating plagues. These results beg the question: How long has Y. pestis been infecting humans? The phylogenic trees generated from recent studies suggested Y. pestis has been with humans for as little as 2,600 years and as long as and 28,000 years. Equipped with these DNA-based tools, Rasmussen et al. asked if they could find evidence of Y. pestis in older human remains.
Continue reading “Yersinia pestis Reveals More Secrets From the Grave”About the Development of an Improved BRET Assay: NanoBRET
One of the more exciting reporter molecules technologies available came online in the past year, with the launch of the Promega NanoBRET™ technology. While it’s easy for me, a science writer at Promega, to brag, seriously, this is a very cool protein interactions tool.
A few of the challenges facing protein-protein interactions researchers include:
- The ability to quantitatively characterize protein-protein interactions
- Ability to examine protein-protein interactions in situ, in the context of the living cell
A goal of the NanoBRET™ developers was to improve the sensitivity and dynamic range of traditional BRET technology, in order to address these challenges.
In May 2015 these researchers published an article outlining their efforts to create NanoBRET technology in ACS Chemical Biology, in an article entitled, “NanoBRET—A Novel BRET Platform for the Analysis of Protein-Protein Interactions”. Here is a brief look at their work.
Continue reading “About the Development of an Improved BRET Assay: NanoBRET”
A Potential Single-Tube Multiplex Assay for Detecting Dengue Virus in the Field
In areas of the world where the electricity is intermittent, resources are limited and transporting bulky equipment and reagents that are sensitive to temperature fluctuations is difficult, diagnosis of viruses like dengue can be challenging. If you could reduce or eliminate the need for electricity dependent equipment for diagnostic assays without sacrificing sensitivity or specificity, it would be a boon to field workers. An article published in PLOS ONE describes how researchers developed a multiplex isothermal amplification method that could assess a potential dengue infection with a visual real-time or endpoint detection in a single tube.