Brachylophosaurus was a mid-sized member of the hadrosaurid family of dinosaurs living about 78 million years ago, and is known from several skeletons and bonebed material from the Judith River Formation of Montana and the Oldman Formation of Alberta. Recent fossil evidence indicates structures similar to blood vessels in location and morphology, have been recovered after demineralization of multiple dinosaur cortical bone fragments from multiple specimens, some of which are as old as 80 Ma. These structures were hypothesized to be either endogenous to the bone (i.e., of vascular origin) or the result of biofilm colonizing the empty network after degradation of original organic components (i.e., bacterial, slime mold or fungal in origin). Cleland et al. (1) tested the hypothesis that these structures are endogenous and thus retain proteins in common with extant archosaur blood vessels that can be detected with high-resolution mass spectrometry and confirmed by immunofluorescence.
Continue reading “Dino Protein: New Methods for Old (Very) Samples”In the Literature
Blogs about published peer-reviewed scientific studies.
Unexpected connections: Gut bacteria influence immunotherapy outcomes
Over the last few years, human microbiome studies have revealed fascinating connections between our colonizing microorganisms and ourselves—including associations between gut bacterial populations and obesity, disease susceptibility, and even mood. The relationship between us and our microbial colonists—once considered completely benign, is now being revealed as an intricate, complicated partnership with the potential to redefine who “we” are in fundamental ways.
Two papers published back-to-back in the November 27 issue of Science add further to this growing body of knowledge—reporting a new and unexpected connection between gut bacterial species and the effectiveness of cancer immunotherapies in mice. The work suggests one reason why such treatments are effective in some circumstances, but not others. Both papers report that the presence of specific bacterial populations may be required for the efficacy of certain treatments, and raise the intriguing question “Could the composition of bacteria in the gut be manipulated to enhance the effectiveness of cancer treatments?”
Continue reading “Unexpected connections: Gut bacteria influence immunotherapy outcomes”ProteaseMAX: A Surfactant for the Most Complex Mixtures
Here we provide two examples of “atypical” experiments that take advantage of the properties of the ProteaseMAX™ Surfactant to improve studies involving digestion of complex protein mixtures.
Example 1
Clostridium difficile spores are considered the morphotype of infection, transmission and persistence of C. difficile infections. A recent publication (1) illustrated a novel strategy using three different approaches to identify proteins of the exosporium layer of C. difficile spores and complements previous proteomic studies on the entire C. difficile spores.
Do you want to build a snowman? Developing and optimizing a qPCR assay to detect ice-nucleating activity
Over the last few months we have published several blogs about qPCR—from basic pointers on avoiding contamination in these sensitive reactions to a collection of tips for successful qPCR. Today we look in depth at a paper that describes the design and and optimization of a qPCR assay, and in keeping with the season of winter in the Northern hemisphere, it is only fitting that the assay tests for the abundance and identity of ice-nucleating bacteria.
Ice-nucleating bacteria are gram-negative bacteria that occur in the environment and are able to “catalyze” the formation ice crystals at warmer temperatures because of the expression of specific, ice-nucleating proteins on their outer membrane. Ice-nucleating bacteria are found in abundance on crop plants, especially grains, and are estimated to cause one-billion dollars in crop damage from frost in the United States alone.
In addition to their abundance on crop plants, ice-nucleating bacteria are also found on natural vegetation and have been isolated from soil, snow, hail, cloud water, in the air above crops under dry conditions and during rain fall. They have even been isolated from soil, seedlings and snow in remote locations in Antarctica. For the bacteria, ice nucleation may be a method to promote dissemination through rain and snow.
Although ice-nucleating bacteria have been isolated from clouds, ice and rain, little is known about their true contribution to precipitation or other events such as glaciation. Are such bacteria the only source of warm-temperature (above temperatures at which ice crystals form without a catalyst) ice nucleation? Can they trigger precipitation directly? What are the factors that trigger their release from vegetation into the atmosphere? Can we determine their abundance and variety in the environment?
Continue reading “Do you want to build a snowman? Developing and optimizing a qPCR assay to detect ice-nucleating activity”A Better GTPase Assay for Drug Development
 The path to drug development is strewn with obstacles: Identifying targets; configuring assays to help identify targets or drugs; uncovering the right compound to affect the selected target without off-target effects and screening multiple compounds to eliminate or identify potential drugs. Without the right tools, compounds or target, identifying potential disease therapies becomes nearly impossible.
The path to drug development is strewn with obstacles: Identifying targets; configuring assays to help identify targets or drugs; uncovering the right compound to affect the selected target without off-target effects and screening multiple compounds to eliminate or identify potential drugs. Without the right tools, compounds or target, identifying potential disease therapies becomes nearly impossible.
When it comes to a drug target for cancer, the Ras protein family is at the top of the list because the proteins are expressed ubiquitiously and found mutated in many types of cancer. Because Ras proteins are involved in transducing signals from the surface of cells, many of the resulting mutations produce an activated Ras, inducing uncontrolled expression of the genes that Ras controls. Ras proteins are small GTPases (20–25kDa) that comprise a larger superfamily of proteins divided into five subfamilies: Ras, Rho, Rab, Arf, and Ran. These proteins control diverse cellular activities, including cellular differentiation, proliferation, cell division, nuclear import and export, and vesicle transport. GTPases are guanosine-nucleotide-binding proteins with affinity for GDP or GTP and are able to hydrolyze GTP. When bound to GTP, GTPases are active (turned on) and interact with downstream proteins in the signaling cascade. When GTPases are bound to GDP, the proteins are inactivated (turned off) and no longer transduce signals. Continue reading “A Better GTPase Assay for Drug Development”
Will Warmer Weather Wake the Sleeping Giant (Viruses)?
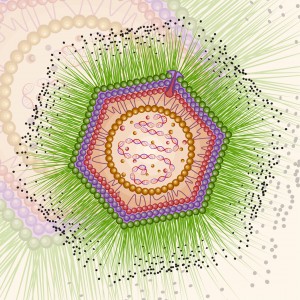
Following the discovery of Mimivirus (1) the first virus with a particles large enough to be visible under the light microscope, two additional “giant” viruses infecting Acanthamoeba have been discovered Pandoravirus (2) and Pithovirus sibericum (3), the latter from a 30,000 year old Siberian permafrost. A fourth type was recently isolated from the same sample of permafrost by Legendre et al, and named Mollivirus sibericum (4).
Mollivirus sibericum has an approximately spherical virion (0.6 µm diameter) with a 651kb GC-rich genome that encodes 523 proteins. To further characterize the virus the researchers performed transcromic- and proteomic-based time course experiments.
For the particle proteome and infectious cycle analysis, proteins were extracted and then run a 4–12% polyacrylamide gel, and trypsin digests were performed in-gel before nano LC-MS/MS analysis of the resulting peptides. Proteomic studies of the particle showed that it lacked an embarked transcription apparatus, but revealed an unusual presence of many ribosomal and ribosome-related proteins.
When the researchers explored the proteome during the course of an entire infectious cycle, the relative proportions of Mollivirus-, mitochondrion-, and Acanthamoeba encoded proteins were found to vary consistently with an infectious pattern that preserved the cellular host integrity as long as possible and with the release of newly formed virus particles through exocytosis.
In an interesting footnote, the authors of this study point out the fact that two different viruses retain their infectivity in prehistorical permafrost layers should be a concern in the context of global warming and the potential to expose humans to primeval viruses.
References
1. La Scola, B. et al. (2003) A giant virus in amoebae. Science 299, 2033.
2. Philippe, N. et al. (2013) Pandoraviruses. Amoeba virus with genomes up to 2.5Mb reaching that of parasitic eukaryotes. Science 341,281–6.
3. Legendre, M. et al. (2014) Thirty thousand year old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc.Natl. Acad. Sci. 111, 4274–9.
4. Legendre, M. et al. (2015) In depth study of Mollivirus sibercum, a new 30,000 year old giant virus infecting Acanthamoeba. Proc. Natl. Acad. Sci. 112, E5327–35 (online).
How Fruit Flies (and maybe Pigeons?) Navigate; A New Report
Several years ago an intriguing story of successful navigation in complex situation, by pigeons, the birds most often compared to rats, caught my eye.
Our backyard once had a coop full of pigeons, so I’m not a total stranger to their navigation abilities (nor am I a pigeon expert). My favorites were the tumbling pigeons.
But it didn’t take much time researching that article from 2012, to learn that one of the more hotly debated how-do-they-do-it topics is animal navigation, in particular, the ability of pigeons to navigate back to home/point A when released at point B.
So when it appeared online today, in Nature Materials, the story “A Magnetic Protein Biocompass” caught my eye.
Continue reading “How Fruit Flies (and maybe Pigeons?) Navigate; A New Report”The Promise of miRNAs as Therapeutic Agents in Treating Disease
When researchers first identified a new family of seemingly non-functional “junk” RNA molecules, it’s unlikely they could have predicted the power and promise of these nucleic acids. The small, non-coding, single-stranded RNAs – typically 21-25 base pairs in length – were first discovered over 20 years ago in C. elegans, yet they were quickly found to be ubiquitous in species from worms to flies to plants to mammals. The role of these novel RNAs in the regulation of developmental pathways in worms, coupled with their prevalence, inspired researchers to better understand their significance.
We now know that miRNAs (for microRNAs) serve as post-transcriptional repressors of gene expression by targeting degradation of mRNA or interfering with mRNA translation. While small, each can have a big effect; a single miRNA can regulate dozens to hundreds of distinct target genes. They’ve been implicated in a variety of critical cellular processes such as differentiation, development, metabolism, signal transduction, apoptosis and proliferation.
Tissue-specific expression patterns revealed that specific miRNAs are enriched in mammalian tissues including adult brain, lung, spleen, liver, kidney and heart. More compelling was the identification of abnormal miRNA expression in tumorigenic cell lines. It’s no wonder that this growing family quickly became ripe for exploration in disease development.

Within only a few years, a rapidly expanding body of research supported the theory that miRNA expression may indeed play a role in the development of human diseases including cardiovascular disease, cancer, diabetes, cystic fibrosis, and liver disease. Investigations into the expression of miRNAs in cardiovascular disease, in particular, have demonstrated not only their value as disease markers, but also how their dysregulation is linked to disease processes.
More recently a new possibility is being explored: can miRNA be manipulated to interfere with disease progression? Continue reading “The Promise of miRNAs as Therapeutic Agents in Treating Disease”
Yersinia pestis Reveals More Secrets From the Grave
Fridays are generally reserved for fun posts to share prior to the weekend. As we all know, fun is relative and to me, the latest news about how long Yersinia pestis has been entwined with human history is intriguing. I enjoy writing about the latest historical finding of Y. pestis even if I do earn a black reputation among my blogging colleagues (pun intended). Therefore, as soon as I saw the Cell article about Y. pestis found in Bronze age human teeth, I knew my blog topic was at hand.
Y. pestis has long been suspected in several plagues that occurred in the last two millennia. Publications in 2011 and 2013 used DNA extracted from teeth of human remains dated to the 14th century Black Death and 6th century Plague of Justinian to confirm Y. pestis was the causative agent in those devastating plagues. These results beg the question: How long has Y. pestis been infecting humans? The phylogenic trees generated from recent studies suggested Y. pestis has been with humans for as little as 2,600 years and as long as and 28,000 years. Equipped with these DNA-based tools, Rasmussen et al. asked if they could find evidence of Y. pestis in older human remains.
Continue reading “Yersinia pestis Reveals More Secrets From the Grave”About the Development of an Improved BRET Assay: NanoBRET
One of the more exciting reporter molecules technologies available came online in the past year, with the launch of the Promega NanoBRET™ technology. While it’s easy for me, a science writer at Promega, to brag, seriously, this is a very cool protein interactions tool.
A few of the challenges facing protein-protein interactions researchers include:
- The ability to quantitatively characterize protein-protein interactions
- Ability to examine protein-protein interactions in situ, in the context of the living cell
A goal of the NanoBRET™ developers was to improve the sensitivity and dynamic range of traditional BRET technology, in order to address these challenges.
In May 2015 these researchers published an article outlining their efforts to create NanoBRET technology in ACS Chemical Biology, in an article entitled, “NanoBRET—A Novel BRET Platform for the Analysis of Protein-Protein Interactions”. Here is a brief look at their work.
Continue reading “About the Development of an Improved BRET Assay: NanoBRET”