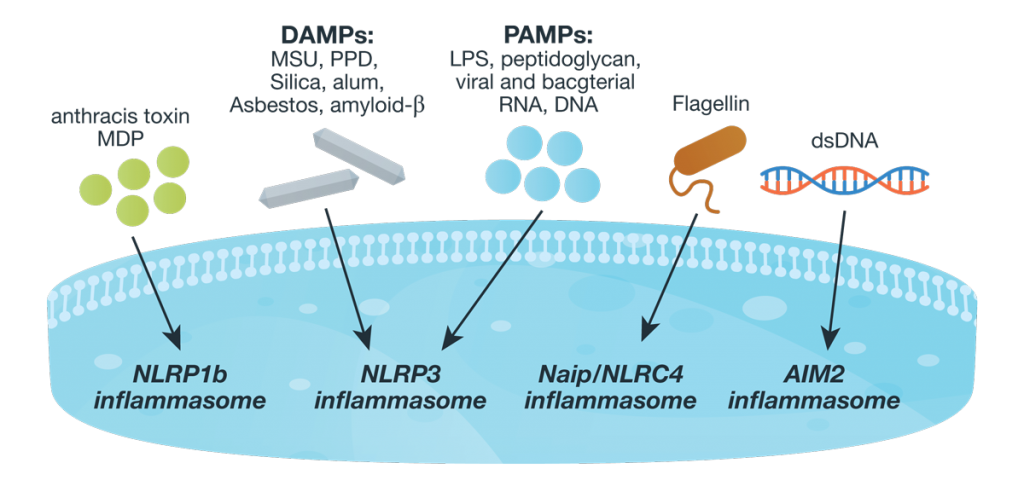
Introduction to Glucose Metabolism

Because glucose metabolism is central to cellular functioning, changes that decrease glucose uptake or increase glycolysis have a widespread effect on on both the cells and organism. How does a simple sugar molecule create such broad effects on health? For example, diabetes results from the inability to store glucose because of a lack of insulin, a hormone that draws glucose from the blood and stores it as glycogen in the liver, muscles and adipose tissue. High levels of sugar in the blood negatively affect the body over the long term, damaging blood vessels and eyesight, making the kidneys work harder to excrete the excess sugar and increasing the risk of stroke and coronary artery disease. Because cancer cells have such a high metabolic demand for glucose, many of the mutations in cancers affect pathways that regulate glucose uptake and glucose breakdown, allowing the cancer cells to survive and grow, crowding out nearby normal cells.
Glucose metabolism is altered by processes other than mutations or an reduced production of a hormone. Throughout its life cycle, a cell will vary its requirements for glucose. For example, the cells that comprise our innate immune response are typically in a quiescent or steady state. However, when these immune cells encounter an foreign invader, they become activated and increase their demand for glucose. To respond to a potential pathogen, the activated cells need glucose to fuel cell proliferation and the production of cytokines, chemicals that activate other immune cells and initiate an inflammatory response. The typical signs of inflammation are red inflamed area that may be painful to the touch, such as a cut that becomes infected. Most inflammation resolves when the infection is eliminated, leaving behind whole skin in the instance of a cut, and the activated immune cells become quiescent again.
An Interesting Observation about Glucose Metabolism in M2 Macrophages
Glucose uptake, immunity and metabolism are cellular pathways that are intertwined such that understanding how glucose is utilized in macrophages illuminates gene induction and regulation in activated macrophages. In a recently published eLife article, Covarrubias et al. studied how activation of murine bone marrow-derived macrophages (BMDMs) by interleukin-4 (IL-4), a signaling cytokine, altered glucose metabolism in the cells and regulated a subset of genes involved in macrophage activation. Continue reading “Finding a Connection Between Glucose Metabolism and Macrophage Activation”

![Fig 4. Four point MMOA screen for tideglusib and GW8510. Time dependent inhibition was evaluated by preincubation of TbGSK3β with 60 nM tideglusib and 6 nM GW-8510 with 10μM and 100μM ATP. (A). Tideglusib [60 nM] in 10μM ATP. (B). GW8510 [60 nM] in 10μM ATP. (C.) Tideglusib [60 nM] at 100μM ATP. (D.) GW8510 [60 nM] at 100μM ATP. All reactions preincubated or not preincubated with TbGSK3β for 30 min at room temperature. Experiments run with 10μM GSM peptide, 10μM ATP, and buffer. Minute preincubation (30 min) was preincubated with inhibitor, TbGSK3β, GSM peptide, and buffer. ATP was mixed to initiate reaction. No preincubation contained inhibitor, GSM peptide, ATP, and buffer. The reaction was initiated with TbGSK3β. Reactions were run at room temperature for 5 min and stopped at 80°C. ADP formed was measured by ADP-Glo kit. Values are mean +/- standard error. N = 3 for each experiment and experiments were run in duplicates. Control reactions contained DMSO and background was determined using a zero time incubation and subtracted from all reactions. Black = 30 min preincubation Grey = No preincubation.](https://www.promegaconnections.com/wp-content/uploads/2016/04/journal.pntd_.0004506.g004-243x300.png)

![One tired pony. By Rachel C from Scotland (Flickr) [CC BY 2.0 (http://creativecommons.org/licenses/by/2.0)], via Wikimedia Commons](https://www.promegaconnections.com/wp-content/uploads/2016/02/Equine_joke-269x300.jpg)