Understanding how a compound or drug affects cellular pathways often requires measuring kinetic changes over an extended period of time—from several hours to days. Live-cell kinetic cell-based assays that measure cell viability, cytotoxicity, apoptosis and other cellular pathways are great for collecting real-time data. You don’t necessarily need expensive equipment to run these types of assays. In the videos below, Dr. Sarah Mahan, a research scientist at Promega, demonstrates how you can easily get great 24-hour or multi-day kinetic data using a GloMax® Microplate Reader.
Continue reading “How to Get Real-Time Kinetic Data With GloMax® Microplate Readers”Bioluminescence
There’s a Microbiome In My Tank!
Imagine a scenario—you’re studying the developmental biology of a species of squid. The squid don’t reproduce in captivity, so females carrying fertilized eggs are collected from the wild and rehomed in your lab’s aquariums. You’ve monitored all the normal aquarium conditions—pH, temperature, salinity—ensuring the animal’s new home mimics its natural environment.
But then, for no reason apparent to you, the clutch of eggs doesn’t develop and doesn’t hatch, derailing your research program until next year when you can collect more adult squid from the wild. What went wrong?

Virus-Like Particles: All the Bark, None of the Bite
Globally, there have been over 5 million deaths attributed to COVID-19 since the start of the pandemic. Throughout the ongoing battle against SARS-CoV-2, researchers have been studying the viral lineage and the variants that are emerging as the virus evolves over time. The more opportunities that the virus has to replicate (i.e., the more people it infects), the greater the likelihood that a new variant will emerge.
The US Centers for Disease Control and Prevention (CDC) classify SARS-CoV-2 variants into four groups: Variants Being Monitored (VBM), Variants of Interest (VOI), Variants of Concern (VOC) and Variants of High Consequence (VOHC). So far, no variants in the US have been identified as VOHC or VOI. Currently, the most common variant in the US is the Delta variant (which includes the B.1.617.2 and AY viral lineages), and it is classified as a VOC.
The Delta variant originated in India and spread rapidly across the UK before making its way into the US (1). Current vaccines, including mRNA and adenoviral vector vaccines, have demonstrated effectiveness against the Delta variant. However, it is a VOC because it is more than twice as contagious as previous variants, and some studies have shown that it is associated with more severe symptoms.
A recent study (2) provides one explanation for the higher infectivity of the Delta variant, using an approach based on virus-like particles (VLPs). The research team was led by Dr. Jennifer Doudna, 2020 Nobel Prize winner for her work on CRISPR-Cas9 gene editing, and Dr. Melanie Ott, director of the Gladstone Institute of Virology at the University of California–Berkeley.
Continue reading “Virus-Like Particles: All the Bark, None of the Bite”Illuminating the Kinome: NanoBRET Target Engagement Technology in the Spotlight
Updated November 21, 2023
In the life of a cell, phosphorylation of proteins is an everyday occurrence. The transfer of a phosphate group, from a molecule such as adenosine triphosphate (ATP) to a specific functional group on a protein, is catalyzed by a protein kinase. The vast majority of protein kinases are classified as either serine/threonine kinases or tyrosine kinases; over 500 kinase genes have been identified in the human genome (1).
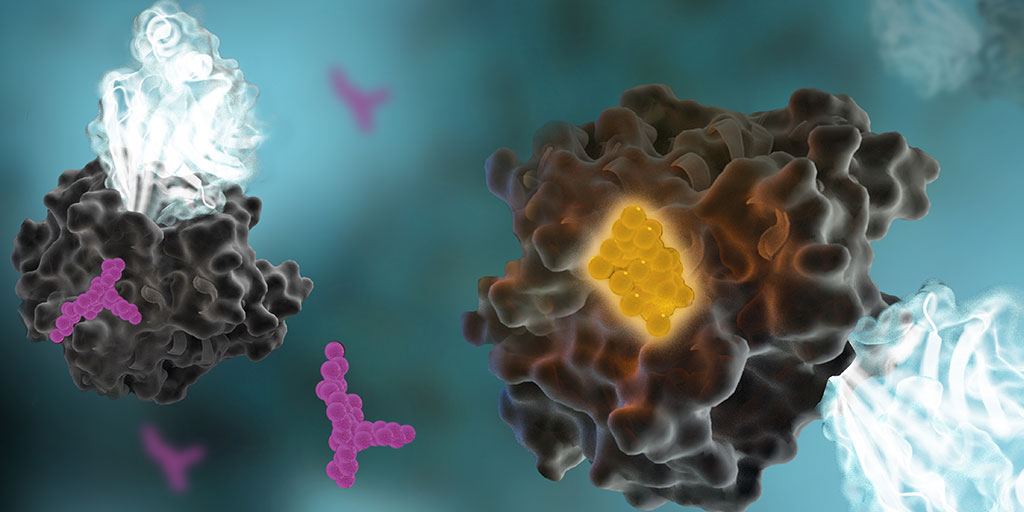
Protein phosphorylation is a key step in most cell signaling pathways, in response to external or internal stimuli, and it is not surprising that dysregulation of these pathways contributes to a variety of cancers. The first oncogene to be characterized was SRC, a gene that encodes a tyrosine kinase (reviewed in 2). With more kinases being implicated in oncogenic pathways, significant drug discovery efforts have been devoted to developing and characterizing inhibitors of protein kinases. These efforts have accelerated ever since the first targeted small-molecule kinase inhibitor, imatinib, received US FDA approval in 2001 for the treatment of chronic myeloid leukemia (3). Since that time, many more protein kinase inhibitors have received FDA approval, with 67 small-molecule inhibitors listed as of September 2021.
Continue reading “Illuminating the Kinome: NanoBRET Target Engagement Technology in the Spotlight”Seeing is Believing: How NanoLuc® Luciferase Illuminates Virus Infections
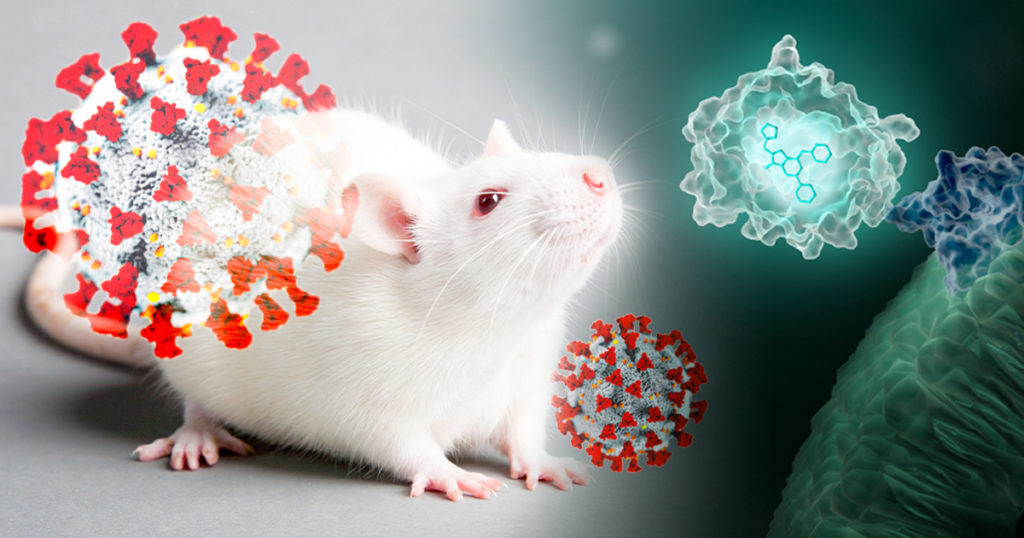
Wearing blue surgical gowns and white respirator hoods, research scientist Pradeep Uchil and post-doctoral fellow Irfan Ullah carry an anesthetized mouse to the lab’s imaging unit. Two days ago, the mouse was infected with a SARS-CoV-2 virus engineered to produce a bioluminescent protein. After an injection of a bioluminescence substrate, a blue glow starts to emanate from within the mouse’s nasal cavity and chest, visible to the imaging unit’s camera and Uchil’s eyes.
“We were never able to see this kind of signal with retrovirus infections.” Uchil is a research scientist at the Yale School of Medicine whose work focuses on the in vivo imaging of retroviral infections. Normally, the mouse would have to be sacrificed and “opened up” for viral bioluminescent signals from internal tissues to be imaged directly.
Continue reading “Seeing is Believing: How NanoLuc® Luciferase Illuminates Virus Infections”LncRNA: The Long and Short of “Junk RNA”
The Central Dogma and Junk DNA

On September 19, 1957, Francis Crick delivered a lecture during a symposium at University College London, titled “Protein Synthesis”. The lecture was published a year later (1); in it, Crick quotes his colleague James Watson as saying, “The most significant thing about the nucleic acids is that we don’t know what they can do.” In contrast, Crick argued that proteins play a central, indispensable role as enzymes within the cell that catalyze a variety of chemical reactions. He believed that the main role of genetic material was to control the synthesis of proteins, although the mechanism of that process was not known.
Crick’s hypothesis came to be known as the central dogma of molecular biology, and it was immortalized in his hand-written notes that described the flow of information from DNA to RNA to proteins. This achievement was all the more remarkable, considering that messenger RNAs were completely unknown at that time, and very little was known about how the cellular translational machinery functioned within the cytoplasm to synthesize proteins. Although the later discovery of retroviruses appeared to challenge Crick’s central dogma, he explained quite succinctly that his original statement had simply been misunderstood, and that information could flow in both directions between DNA and RNA (2).
Continue reading “LncRNA: The Long and Short of “Junk RNA””Permeability Possibilities with the New NanoClick Assay
Peptide Predicament
For decades now, peptides have been a molecule of interest for drug discovery research. Peptides offer a unique opportunity for therapeutic intervention that closely mimics natural pathways, as many physiological functions utilize peptides as intrinsic signaling molecules. Macrocyclic peptides, in particular, have recently proven to be promising candidates for targeting intracellular protein–protein interactions (PPIs), an attractive but hard-to-reach therapeutic target for conventional small molecule and biological drugs.
As with any opportunity, there are also challenges that accompany the peptide therapeutic development. Peptide ligands typically have poor membrane permeability, so thus far the majority of peptide therapeutics predominantly target extracellular proteins and receptors. There are also multiple mechanisms for cellular uptake of peptides, including both energy-dependent routes like endocytosis, and energy-independent, like passive diffusion or membrane translocation. Multiple mechanisms of cellular uptake paired with poor permeability makes engineering enough membrane permeability into peptides in order to advance them through drug discovery pipelines extremely difficult.
There are other factors to consider in developing peptide therapeutics, such as solubility, protein/lipid binding and stability, which can also have an affect on the overall cytosolic concentration and, ultimately impact the ability of the peptide to effectively engage its desired intracellular targets.
With so many challenging factors, the ability to have a predictive, high-throughput assay to assess cell permeability, independent of the mechanism(s) of entry, would be a critical and invaluable tool to support peptide drug discovery research.
In a recent study published in ACS Chemical Biology, researchers sought to develop such a tool, and demonstrated a new application for Promega NanoBRET™ technology: the NanoClick assay.
Continue reading “Permeability Possibilities with the New NanoClick Assay”World Firefly Day: Shining More Light on Glo-ing Innovations
On July 3rd and 4th, 2021, we celebrate World Firefly Day, and 2021, marks 30 years of luciferase products firefly luciferase vectors and Luciferase Assay System. These tools are key in advancing bioluminescent technology. To celebrate this day, we want to highlight some innovations that have been made possible with these tools.
Continue reading “World Firefly Day: Shining More Light on Glo-ing Innovations”Bringing Cutting Edge Technologies to Academic Researchers Through the Academic Access Program
This post was written by guest blogger Iain Ronald, Director Academic/Government Market Segment at Promega.
My back story is similar to most of you reading this blog, high school education, undergraduate degree then onto a postgraduate degree. However, over 25 years ago during my undergraduate study, I was fortunate enough to work in the lab of Professor Ray Waters studying DNA damage in S. cerevisiae as a model organism and at the time PCR was cutting-edge technology and the PCR license was in full effect. However, there was one company that was fighting the good fight to democratize PCR for the good of the scientific community, Promega.
I became enamored with Promega then, and the next steps in my career were taken with a view to working at this company who, for all intents and purposes, seemed to really care about the progression of science beyond self-aggrandizement.
Now that I am working at Promega in a position where I can bring benefit to our academic community, I have pondered what I can do to equal the disruptive attitude I observed in this company all those years ago when they were fighting the then “big tech” for the enablement of the scientific community.
How Promega Helped Our Lab Scale Up Drug Discovery for Bloodborne Pathogens
This blog was written by Sebastien Smick, Research Technician in Dr. Jacquin Niles’ laboratory at Massachusetts Institute of Technology (MIT)
Our lab is heavily focused on the basic biology and drug discovery of the human bloodborne pathogen Plasmodium falciparum, which causes malaria. We use the CRISPR/Cas9 system, paired with a TetR protein fused to a native translational repressor alongside a Renilla luciferase reporter gene, to conditionally knock down genes of interest to create modified parasites. We can then test all kinds of compounds as potential drug scaffolds against these gene-edited parasites. Our most recent endeavor, one made possible by Promega, is a medium-low throughput robotic screening pipeline which compares conditionally-activated or-repressed parasites against our dose-response drug libraries in a 384-well format. This process has been developed over the past few years and is a major upgrade for our lab in terms of data production. Our researchers are working very hard to generate new modified parasites to test. Our robots and plate readers rarely get a day’s rest!
