The cause of type 1 diabetes (T1D) is not well understood. What is known is that in T1D, immune cells attack pancreatic islet cells that produce insulin. In addition, insulin is an autoantigen that activates T cells in diabetic persons.
A new discovery by Ahmed et al. could further T1D understanding. These findings are also setting B and T cell paradigms on their ear.
About B Cells and T Cells
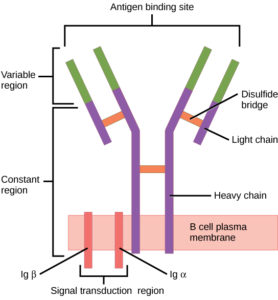
B cells (B lymphocytes) are part of the cellular immune response. They act by means of surface receptor molecules that are immunoglobulins. These B cell receptors are created by highly variable gene rearrangements that result in a huge variety of these surface immunoglobulin molecules. The beauty of B cell receptors (BCR) lies in the fact that, through random gene rearrangements comes a such large variety of B cell surface receptors, that any foreign antigen that makes its way into the body is recognized and snagged by a B cell receptor.
B cells then internalize, process and present these antigens to T cells.
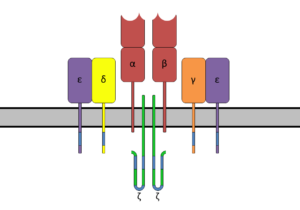
T cells (T lymphocytes) also have unique receptor molecules on their surface. The T cell receptor (TCR) recognizes fragments of antigen bound to major histocompatibility complex (MHC) molecules.
The TCR is composed of the highly variable alpha (α) and beta (β) chains expressed as part of a complex with the invariant CD3 chain molecules. The CD3 T cell coreceptor helps to activate both the cytotoxic T cell (CD8+ T cells) and also T helper cells (CD4+ T cells).
A Unique Lymphocyte: Part B Cell, Part T Cell
Unique lymphocytes expressing both BCR and TCR, were found in peripheral blood of several individuals. Labelled dual expressers (DEs), most of these cells expressed IgD and IgM, with a minor subset expressing IgG and IgA—these appeared to be class switching DEs. The authors sorted the DEs by flow cytometric imaging, noting that DEs were found “significantly more frequent[ly] in T1D subjects than in healthy controls (HCs)”.
Ahmed et al. reported their findings in the journal Cell in May: “A public BCR present in a unique dual-receptor-expressing lymphocyte from type 1 diabetes patients encodes a potent T cell autoantigen“. They believe these dual-expresser cells to be a new type of lymphocyte, as they have not previously been reported. They note that in the T1D patients with these dual-expresser cells (DEs), a single clonotype predominates and this cell type encodes a CD4 T cell autoantigen at its antigen binding site.
Understanding the Dual-Expresser Cells
The authors investigated the dual phenotype of DEs at the single-cell level by examining transcriptomes with single-cell RNA sequencing. They sorted individual DEs and B cell and T cell controls from peripheral blood mononuclear cells (PBMCs) and analyzed the transcriptomes using a SMART-seq2 method. A total of 77 cells passed their quality control standards based on expression of at least 2 of 3 housekeeping genes. Of these cells, it was found that DEs expressed unique genes, as well as genes for both T and B cells.
Next, Ahmed et al. showed shared expression of B and T cells markers at the protein level, using fluorescence-activated cell sorting. They then analyzed DEs for standard components of BCR—CD79a and CD79b0, and TCR—CD3gamma, CD3 epsilon CD3delta and CD247. These receptor components are responsible for activation signals. All components in DEs were found to match those in the B and T cell controls used.
Unique IGHV Regions In DEs
The authors analyzed the IGHV—the hypervariable region of the DEs. They compared IGHV regions of IgD+ and IgD– DEs and B cell controls by cell sorting. These results were then compared to IGHV sequences of B cell controls from each of 3 subjects. Results showed a predominant IGHV04-b gene used in DE cells from three subjects (the gene was used by 95% of IgD+ cells in T1D#1, 22% of IgD– cells in subject #2 and 88% of D+ and D– cells in subject #3).
In contrast, the IGHVH04-b gene was used by less than 1% of B cell controls in the 3 subjects. And they found no significant overlap in VH use by dual-expressors and B cell controls; the authors report that the top 10 VH genes used in B cell controls of the three subjects were either absent in their DEs or appear as only minor components in the VH repertoire in DEs.
The common IGHV04-b in DEs from several subjects was further explored by examining the IGHV04-bs for clonality, which showed the IGHV04-b DEs of the three subjects to be of a single clonotype, with the same VH, DH and JH segments, as well as N1 and N2 junctions. Thus the subjects had CDR3 regions with identical nucleotide and amino acid sequences. The authors named this unique CDR3 the “x-clonotype.”
X-Clonotype in Healthy Controls?
Very few DE cells have been found in healthy controls (HC). To understand the prevalence of the x-clonotype, Ahmed et al. compared IgD+ and IgD– DEs to B cell controls, from a single HC. The DEs and B cell controls all had diverse repertoire. IGHV04-b usage was rare in DEs and B cell controls of the HC. The authors concluded that the x-clonotype was absent from cells of HCs.
Furthermore, their survey of a public database found no x-clonotype from high-resolution immunoSEQ data of 37 million unique BCR sequences, of both naive and memory B cells from healthy subjects.
Ahmed et al. further demonstrated the potential importance of the x-clonotype by clonally expanding DEs and determining that they encode a potent autoantigen in the variable region of the Ig heavy chain. This autoantigen (“x-autoantigen”) has
“an optimal register for the diabetogenic DQ8 molecule.”
Peptides of the x-autoantigen were shown to form complexes with DQ8 molecules that stimulate CD4 T cells from T1D but not HC subjects.
Significance and Future Studies
The authors note that these findings
“lay the groundwork for examining whether the x-autoantigen could serve as a T1D risk biomarker.”
DQ8 is a major risk factor for T1D and is carried by about 90% of T1D patients. But most DQ8 subjects do not develop T1D. In this work, the x-clonotype was seen in DQ8 T1D subjects but not in DQ8 HCs. Additionally, the x-clonotype was not found in HCs with other DQs. The authors propose screening more at-risk subjects to determine the value of the x-clonotype as a biomarker as well as performing more single-cell RNA-seq analysis to determine whether DEs are a distinct, new cell type, or a subpopulation of an existing cell.
The number of human subjects from which this discovery was made is obviously very small but the results are an exciting step towards better understanding of autoimmune disease, particularly T1D. It will be interesting to see future results from these researchers.
Here’s the paper:
Ahmed, et al. (2019) A Public BCR Present in a Unique Dual-Receptor-Expressing Lymphocyte from Type 1 Diabetes Patients Encodes a Potent T Cell Autoantigen. Cell 177(6), 1583–99.
