They existed 3.5 billion years before humans evolved on Earth. They’re neither dead nor alive. Their genetic material is embedded in our own DNA, constituting close to 10% of the human genome. They can attack most forms of life on our planet, from bacteria to plants to animals. And yet, if it wasn’t for them, humans might never have existed.
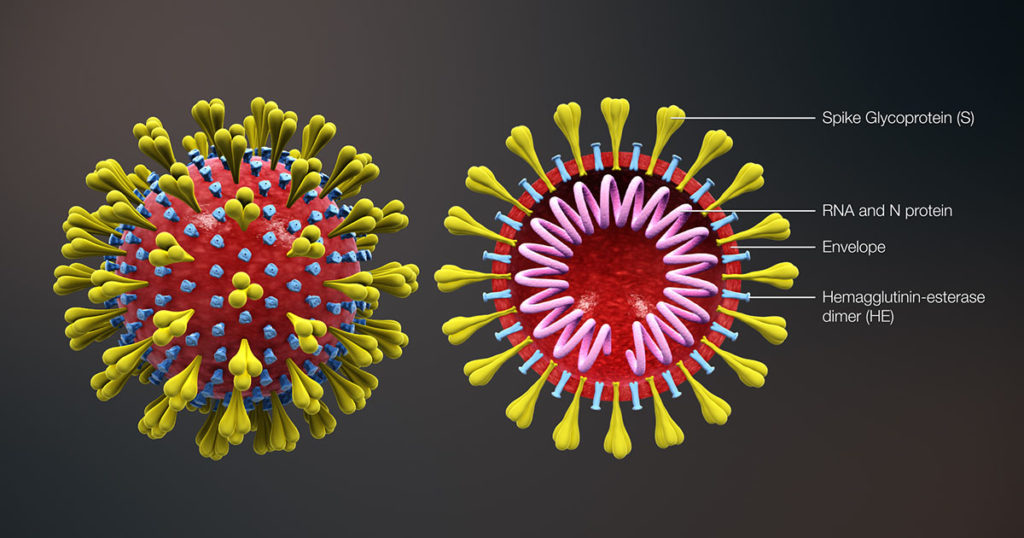
No, that’s not the blurb for a new Hollywood blockbuster, although recent developments have proven, once again, that truth is decidedly more bizarre than fiction. Now that “coronavirus” has become a household word, the level of interest in all things virus-related is growing at an unprecedented rate. At the time of writing, coronavirus and COVID-19 topics dominated search traffic on Google, as well as trends on social media. A recent FAQ on this blog addresses many of the questions we hear on these topics.
Continue reading “Which Came First: The Virus or the Host?”


 GPCRs
GPCRs
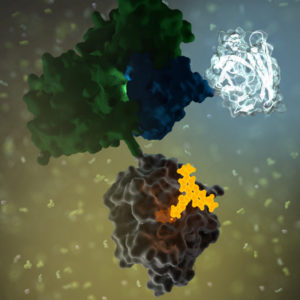
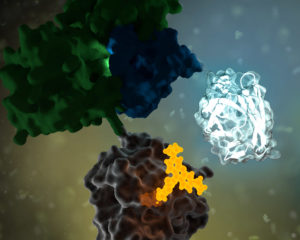 No protein is an island. Within a cell, protein-protein interactions (PPIs) are involved in highly regulated and specific pathways that control gene expression and cell signaling. The disruption of PPIs can lead to a variety of disease states, including cancer.
No protein is an island. Within a cell, protein-protein interactions (PPIs) are involved in highly regulated and specific pathways that control gene expression and cell signaling. The disruption of PPIs can lead to a variety of disease states, including cancer.