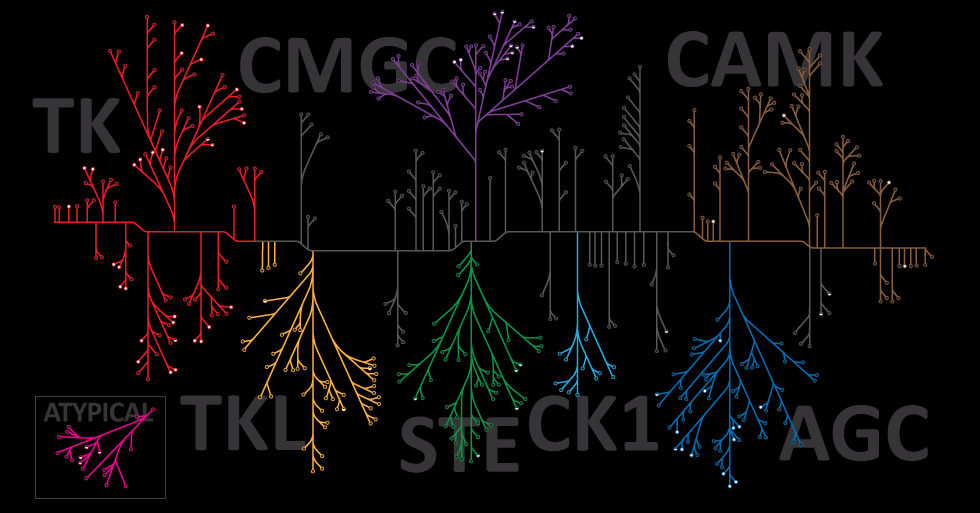
Using Pepsin to Prepare F(ab’)2 Fragments
Pepsin is commonly used in the preparation of F(ab’)2 fragments from antibodies. In some assays, it is preferable to use only the antigen-binding (Fab) portion of the antibody. For these applications, antibodies may be enzymatically digested to produce an F(ab’)2 fragment of the antibody. To produce an F(ab’)2 fragment, IgG is digested with pepsin, which cleaves the heavy chains near the hinge region. One or more of the disulfide bonds that join the heavy chains in the hinge region are preserved, so the two Fab regions of the antibody remain joined together, yielding a divalent molecule (containing two antibody binding sites), hence the designation F(ab’)2. The light chains remain intact and attached to the heavy chain. The Fc fragment is digested into small peptides.
Continue reading “Using Pepsin to Prepare F(ab’)2 Fragments and Determine Deuterium Exchange”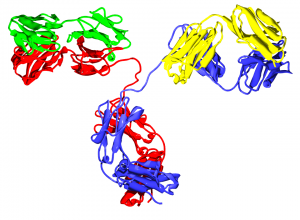
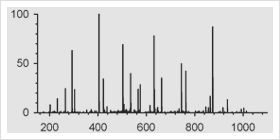



![0877MA10_3A-[Converted]](https://www.promegaconnections.com/wp-content/uploads/2011/06/0877ma10_3a-converted.jpg?w=300)