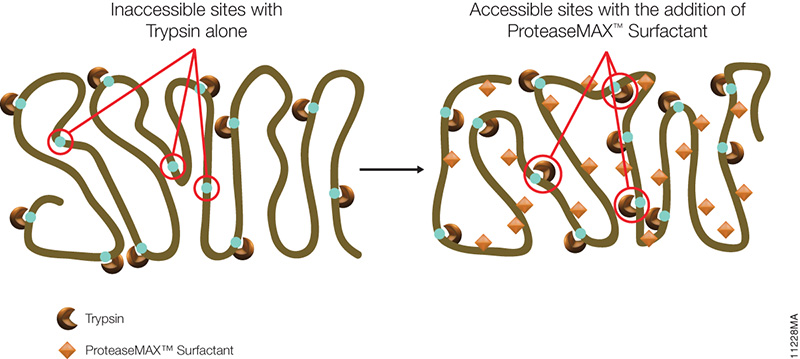
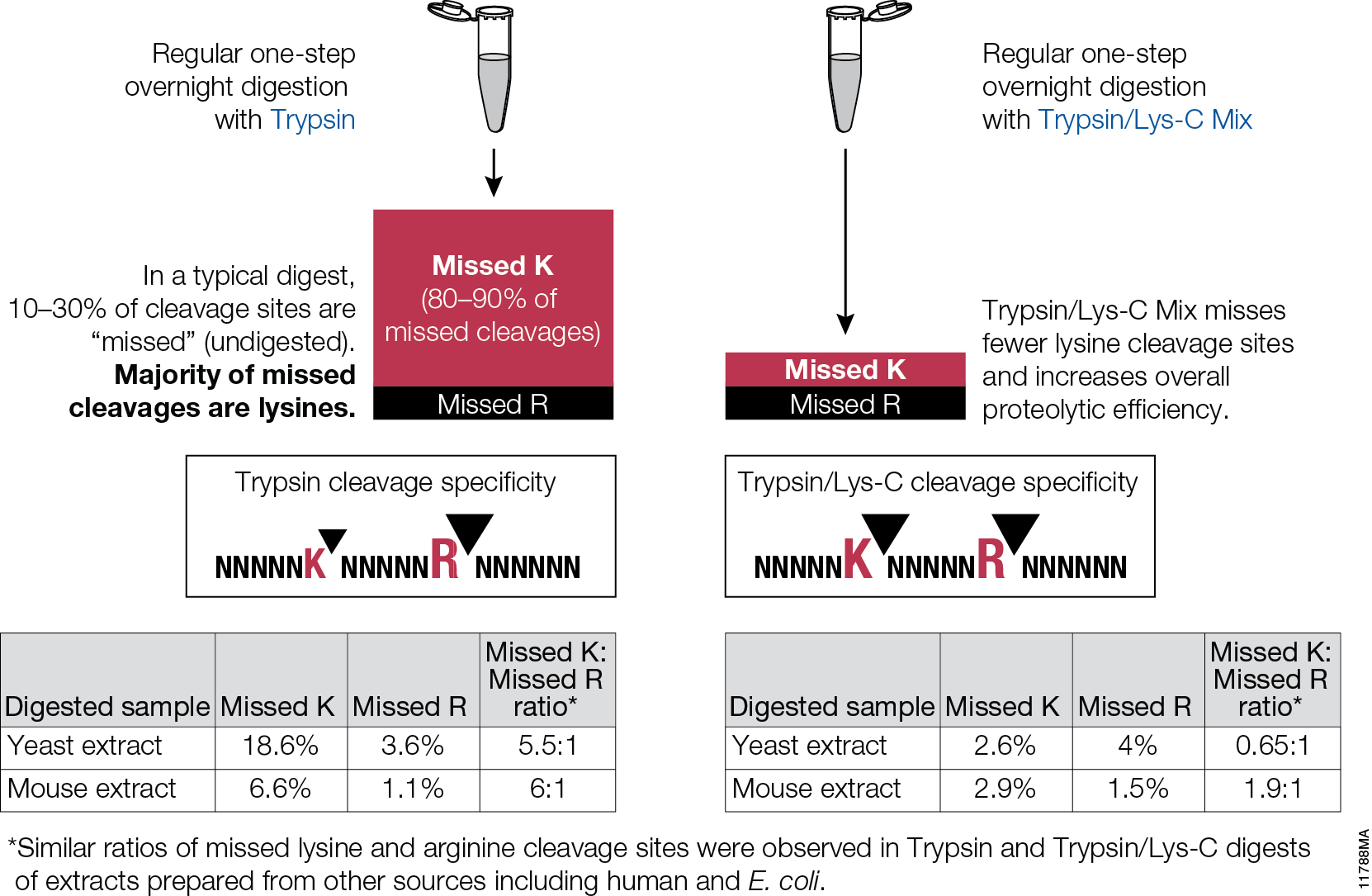
Food allergies are increasing worldwide and becoming a public health issue, especially among children are concerned. Children have a higher prevalence of food allergies, with about 4–8%, compared to adults (1–5%). Currently antibody-based methods such ELISA (enzyme-linked immunosorbent assay) are the primary method for food allergen analysis. In most cases antibodies are only available for single well-known allergens. Often those that are commercially available are poorly characterized resulting cross-reactivity that leads to false-positive results in diagnostic tests.
A recent publication (1) presented a review of an alternative technology based on mass spec (i.e., multiple reaction monitoring, MRM) that circumvents the drawbacks of antibody based methods. MRM allows precise quantitative determination of target proteins in complex samples with broad dynamic range. MRM also provides quantification of different isoforms. It is noted that tryptic digestion followed by mass spec analysis, has already identified several unique peptides for different allergens, including those found in crustaceans, eggs, fish, peanuts, soy and wheat. In summary the challenge is now to select the appropriate tryptic signature peptide(s) for the respective allergen and to develop well characterized standards (i.e., isotope labeled standards) to ensure accurate quanititation.
Citation:
Koeberl, M et al. (2014) Next Generation of Food Allergen Quantification using Mass Spectrometric Systems J. Proteome Research 13, 3499–509.
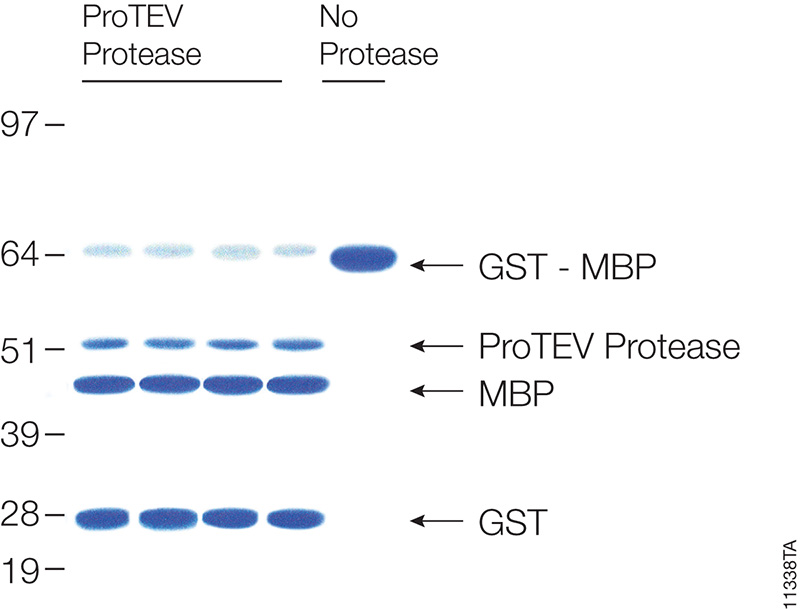
 Many different polypeptide fusion partners or affinity tags have been developed to facilitate purification of target proteins. The most commonly used tag for the purification and detection of recombinant expressed proteins is the His tag. Cloning vectors designed to generate His-tagged proteins contain 5–10 histidine residues at either the C- or N terminus of the expressed protein. The His tag adds only 0.84kDa to the mass of the protein and is nonimmunogenic. Also, because the tertiary structure of the tag is not important for purification, His-tagged proteins can be purified using native or denaturing conditions. The affinity of histidine residues for immobilized nickel allows selective purification of His-tagged proteins. The
Many different polypeptide fusion partners or affinity tags have been developed to facilitate purification of target proteins. The most commonly used tag for the purification and detection of recombinant expressed proteins is the His tag. Cloning vectors designed to generate His-tagged proteins contain 5–10 histidine residues at either the C- or N terminus of the expressed protein. The His tag adds only 0.84kDa to the mass of the protein and is nonimmunogenic. Also, because the tertiary structure of the tag is not important for purification, His-tagged proteins can be purified using native or denaturing conditions. The affinity of histidine residues for immobilized nickel allows selective purification of His-tagged proteins. The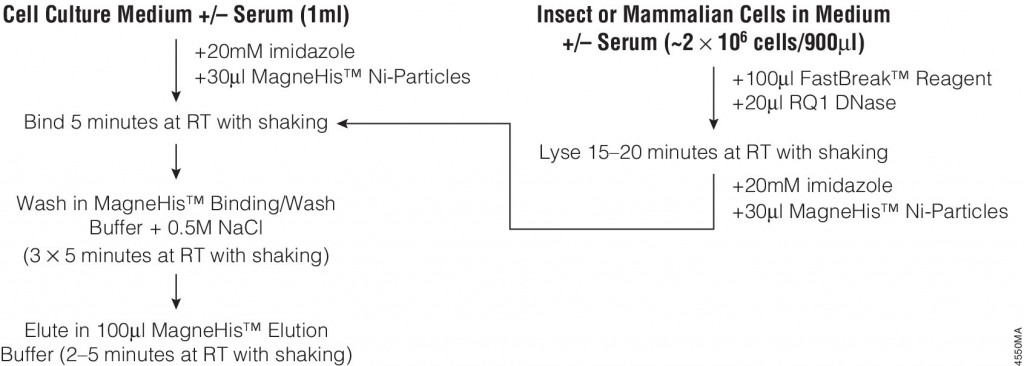
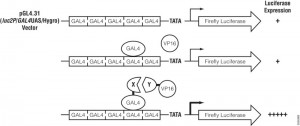
 Although previous references have provided data regarding the potential oncogenic role of the gene ETV7, there has been minimal investigation as to its physiological role.
Although previous references have provided data regarding the potential oncogenic role of the gene ETV7, there has been minimal investigation as to its physiological role.