
Back in the fall, I received a sampling kit, an Informed Consent form and instructions for collecting samples for the Wild Life In Our Homes citizen science project. I carefully swabbed the requested surfaces: exterior and interior door trim, kitchen counter tops, pillowcases, etc., and sent my samples in. I later received confirmation that my samples had been received and again later confirmation that they were being analyzed.
The first paper from this project has been published by Dunn et al. in PLOS ONE (Home Life: Factors Structuring the Bacterial Diversity Found within and between Homes). This initial report covers the first 40 homes sampled, all from the Raleigh-Durham, NC, USA area. Volunteers sampled their homes in the Fall of 2011, collecting specimens from nine areas: cutting boards, kitchen counters, refrigerator, toilet seat, pillowcase, door handle, TV screen, and interior and exterior door trim. The scientists used direct PCR and high-throughput sequencing to sequence the bacterial 16S rRNA gene from the submitted samples. By doing this they were able to estimate the diversity within each sample—they did not distinguish between live and dead organisms, and they did not sequence anything other than the bacterial 16SrRNA, so this study is limited to bacteria.
Among all the samples, they identified 7726 unique phylotypes based on their molecular analyses. On average, they identified 2252 Operational Taxonomic Units (OTUs) for each home sampled. When they looked at all nine samples combined for all the homes, they saw that diversity increased where aerosols and airborne particles were most likely: TV screen and interior and exterior door trims.
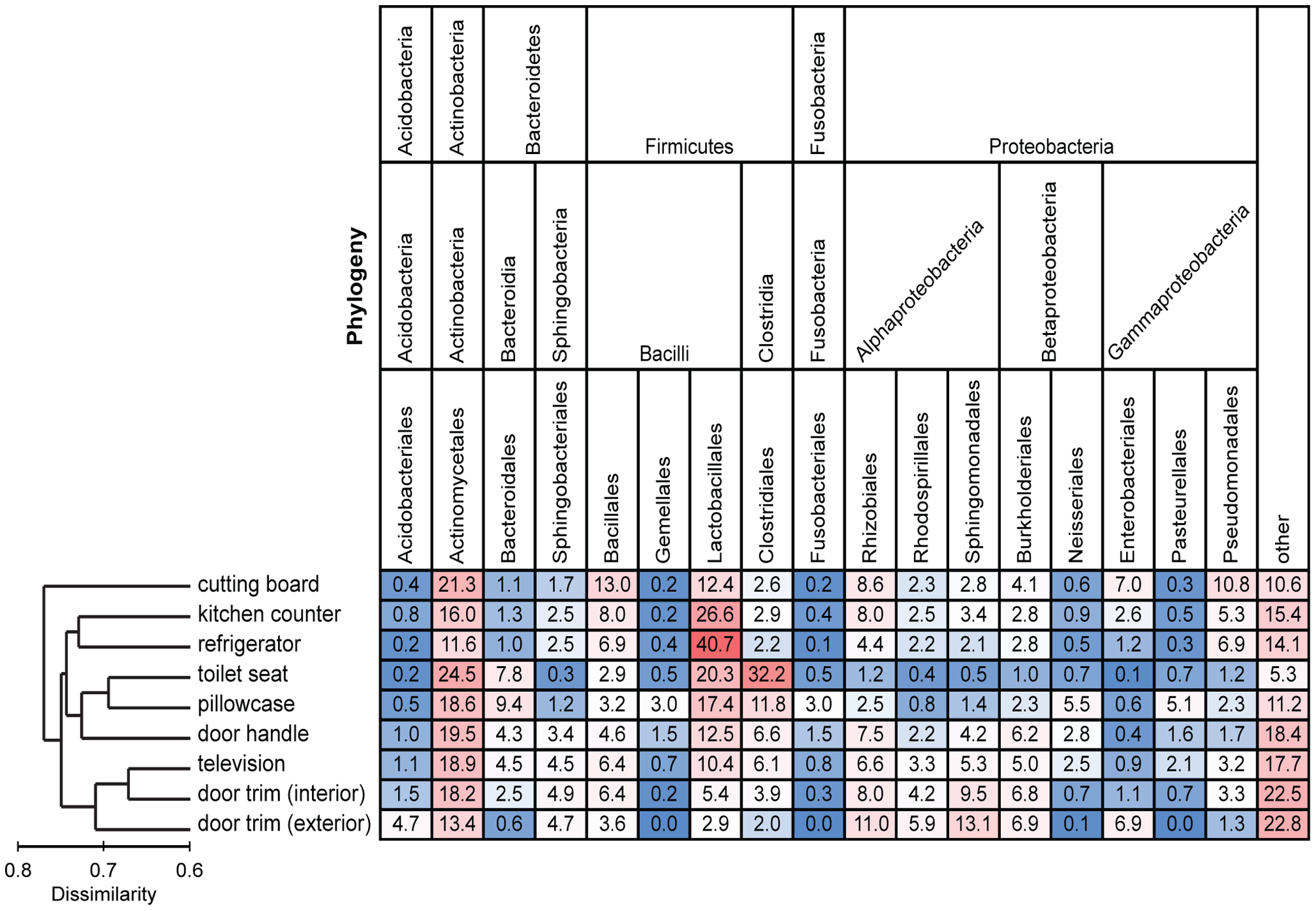
Each column is colored so that taxa with high relative abundances are red, intermediate relative abundances are white, and low abundances are blue. The dendrogram on the left summarizes the overall degree of dissimilarity (calculated from unweighted UniFrac values) of the bacterial communities in each of the sampled locations relative to each other. The dendrogram was created using mean UniFrac values for each of the sampled locations. Figure and legend from Dunn et al. PLOS ONE
doi:10.1371/journal.pone.0064133.g003
The phyla that were most common based on number of sequences obtained were Protobacteria, Fimicutes and Actinobacteria. The bacterial families that were most well represented across all the sample types were Streptococcaceae, Cornyebacteriaceae and Lactobacillaceae.
Each home seemed to have its own special community composition, but they could identify large buckets into which samples clustered as far as community composition. Similar bacteria were found in decompositional environments (door trims, TV screen); kitchen-associated environments and frequently touched surfaces (pillowcases, door handle, toilet seat).
The researchers compared their sample phylotypes to previous research on bacterial sources around the home. As would be expected, human sources, particularly skin, probably contributed to many of the bacteria found on interior surfaces (Streptococcaceae, Corynebacteriaceae, and Lactobacillaceae). Bacteria that are typically associated with leaves and produced were represented on door trim surfaces.
Lifestyle of occupants also affected the wild life found in the homes. For instance, the presence of dogs affected the phylotypes identified from pillowcase and TV screen samples, with Porphyromonadaceae, Rhodocyclaceae and Pasteruellaceae being abundant in these cases (these are common on dog skin and fur). However, the presence of cats did not seem to affect the bacterial communities identified; the authors do state they only have a sample size of three, and this may change as more data are collected. Children, use of pesticides, carpet also did not show significant influence on bacterial communities identified.
This paper is just the beginning, and it probably raises more questions than it answers. For instance, will a larger sample size show an affect of cats or children on bacterial communities? Will the same thing be seen for fungi or other kinds of microorganisms? Will there be any correlation between the type of microbial communities or their diversity and the overall health of the occupants of the home?
But what I really want to know is what is thermophillic organism that is growing in the steam vent of my dishwasher… it’s just a little scary. But they didn’t sample that surface.
Michele Arduengo
Latest posts by Michele Arduengo (see all)
- An Unexpected Role for RNA Methylation in Mitosis Leads to New Understanding of Neurodevelopmental Disorders - March 27, 2025
- Unlocking the Secrets of ADP-Ribosylation with Arg-C Ultra Protease, a Key Enzyme for Studying Ester-Linked Protein Modifications - November 13, 2024
- Exploring the Respiratory Virus Landscape: Pre-Pandemic Data and Pandemic Preparedness - October 29, 2024

Michele, I commend you for participating in a public study such as this, and in posting the results. Would also add that any report that has “pillowcase” and “toilet seat” in the same phrase makes me somewhat uncomfortable, no matter the organism involved. Interesting blog! And if your dishwasher thermophile is orangish-pink, Serratia sp. maybe? Not that I’ve seen such a thing at my place….
Hi Kari,
I hadn’t thought about the juxtaposition of pillowcase and toilet seat, but your point is well taken. And no, it’s not pink, it’s black. I’m actually wondering if it’s a mold of some kind.