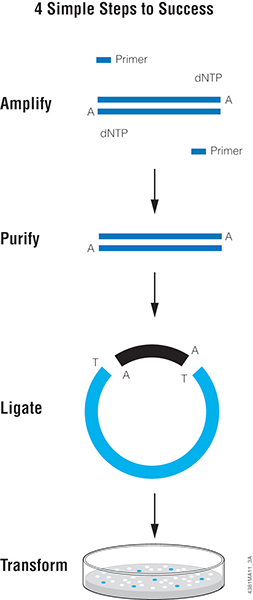
One of the easiest methods for cloning blunt-ended DNA fragments including PCR products is T-vector cloning, such as with pGEM®-T or pGEM®-T Easy Vector Systems. This method takes advantage of the “A” overhang added by a PCR enzyme like Taq DNA Polymerase. T vectors are linearized plasmids that have been treated to add 3′ T overhangs to match the A overhangs of the insert. The insert is directly ligated to the T-tailed plasmid vector with T4 DNA ligase. The insert can then be easily transferred from the T vector to other plasmids using the restriction sites present in the multiple cloning region of the T vector.
Proofreading polymerases like Pfu do not add “A” overhangs so PCR products generated with these polymerases are blunt-ended. In a previous blog, we discussed a simple method for adding an A-tail to any blunt-ended DNA fragment to enable T-vector cloning. Below, we think about the next step: Ligation.
Ligation Preparation
You have blunt-ended your DNA insert of interest (from PCR, restriction enzyme digestion, or even sheared and blunted genomic DNA), made sure the fragment is A tailed and are ready to clone into a T vector (e.g., pGEM®-T Easy Vector). The next step is as simple as mixing a few microliters of your purified product with the cloning vector in the presence of DNA ligase, buffer and ATP.
Estimate the concentration of the prepared insert DNA by spectrophotometric method or compare the staining intensity of your PCR product with that of DNA molecular weight standard of similar size and known concentration on an agarose gel. If the vector DNA concentration is unknown, estimate the vector concentration by the same method you chose for the insert.
It’s a good idea to set up various vector:insert DNA ratios to determine the optimal ratio for a particular vector and insert. Keep in mind, the vector:insert ratio will change depending on the size of the insert. In most cases, a 1:1 or 1:3 molar ratio of vector:insert works well, but you may want to consider 1:5, 5:1 and even a 10:1 ratio.
The equation for calculation of the amount of insert required at a specific molar ratio of vector:insert is:
[(ng of vector × kb size of insert) ÷ kb size of vector] × (molar amount of insert ÷ molar amount of vector) = ng of insert
Example:
How much 500bp insert DNA needs to be added to 100ng of 3.0kb vector in a ligation reaction for a desired vector:insert ratio of 1:3?
Answer: [(100ng vector × 0.5kb insert) ÷ 3.0kb vector] × (3 ÷ 1) = 50ng insert
If the math looks intimidating, don’t worry. Our BioMath Calculators are here to help!
If you’re interested in more details about how to set up a ligation, take a look at this helpful video:
Tips for T-Vector Cloning
Here are a few things to remember while you work:
- Nucleases may degrade the T overhangs on the vector. Make sure you use sterile, nuclease-free water in your ligation reactions.
- Use high-efficiency competent cells (≥1 × 108cfu/μg DNA) for transformations. Ligation of fragments with a single-base overhang can be inefficient, so it’s essential to use cells with a high transformation efficiency to obtain a reasonable number of colonies.
- Limit exposure of your PCR product to shortwave UV light to avoid formation of pyrimidine dimers. Use a glass plate between the gel and UV source. If possible, only visualize the PCR product with a long-wave UV source, or use something like Diamond® Nucleic Acid Dye that allows you to visualize your DNA without the use of UV.
For more information on cloning, consult our Subcloning Technology Guide.
Are you an student or early-career scientist looking for troubleshooting and how-to resources? Or, do you supervise entry-level lab members? Check out our Student Resource Center for more helpful articles, videos and tools!
Related Posts
Latest posts by Leah Cronan (see all)
- Mass Spec for Glycosylation Analysis of SARS-CoV-2 Proteins Implicated in Host-Cell Entry - November 10, 2020
- Proteomics from a Different Point of View: Introducing ProAlanase, the Newest Mass-Spec Grade Protease from Promega - August 7, 2020
- Go fISH! Using in situ Hybridization to Search for Expression of a SARS-CoV-2 Viral Entry Protein - July 10, 2020

if the vector is in linear form then how is it possible for it to produce blue colonies without a insert, the alpha fragment is disrupted if it is in a linear form.
Due to incomplete digestion or re-ligation, there will be a few circular plasmids. That’s why you need to estimate the background colonies.