Synthetic biology—genetically engineering an organism to do or make something useful—is the central goal of the iGEM competition each year. After teams conquer the challenge of cloning their gene, the next hurdle is demonstrating that the engineered gene is expressing the desired protein (and possibly quantifying the level of expression), which they may do using a reporter gene.
Reporters can also play a more significant role in iGEM projects when teams design their organism with reporter genes to detect and signal the presence of specific molecules, like environmental toxins or biomarkers. Three of the iGEM teams Promega sponsored this year opted to incorporate some version of NanoLuc® Luciferase into their projects.
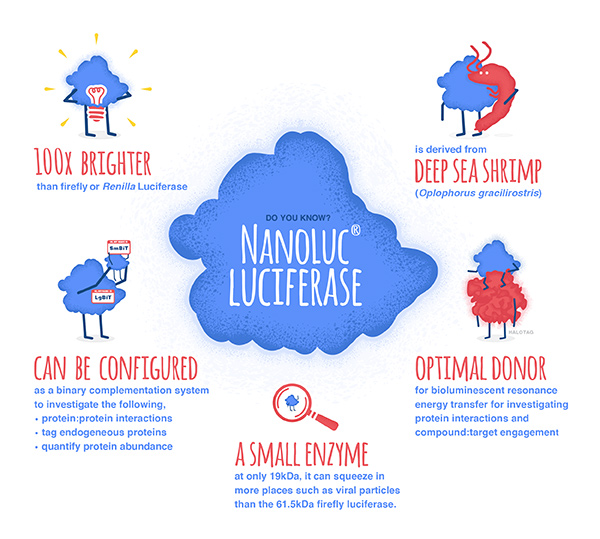
NanoLuc® luciferase is a small monomeric enzyme (19.1kDa, 171 amino acids) based on the luciferase from the deep sea shrimp Oplophorus gracilirostris. This engineered enzyme uses a novel substrate, furimazine, to produce high-intensity, glow-type luminescence in an ATP-independent reaction. Unlike other molecules for tagging and detecting proteins, NanoLuc® luciferase is less likely to interfere with enzyme activity and affect protein production due to its small size.
NanoLuc® Luciferase has also been engineered into a structural complementation reporter system, NanoBiT® Luciferase, that contains a Large subunit (LgBiT) and two small subunit options: low affinity SmBiT and high affinity HiBiT. Together, these NanoLuc® technologies provide a bioluminescent toolbox that was used by the iGEM teams to address a diverse set of biological challenges.
Here is an overview of each team’s project and how they incorporated NanoLuc® technology.
Using NanoLuc® to Create a Synthetic Biosensor
NeuroDrop, the 2019 project of the Grenoble-Alpes team, is a device capable of detecting small amounts of biomarkers from low volume samples, such as tears. The Grenoble-Alpes team focused on the potential for lacrimal fluid to reveal pathophysiological changes in the central nervous system and lead to a non-invasive technique for diagnosing neurodegenerative disorders. In order to enable sensitive biomarker detection from just a few microliters, they proposed developing an innovative synthetic biosensor and a smart hardware device.
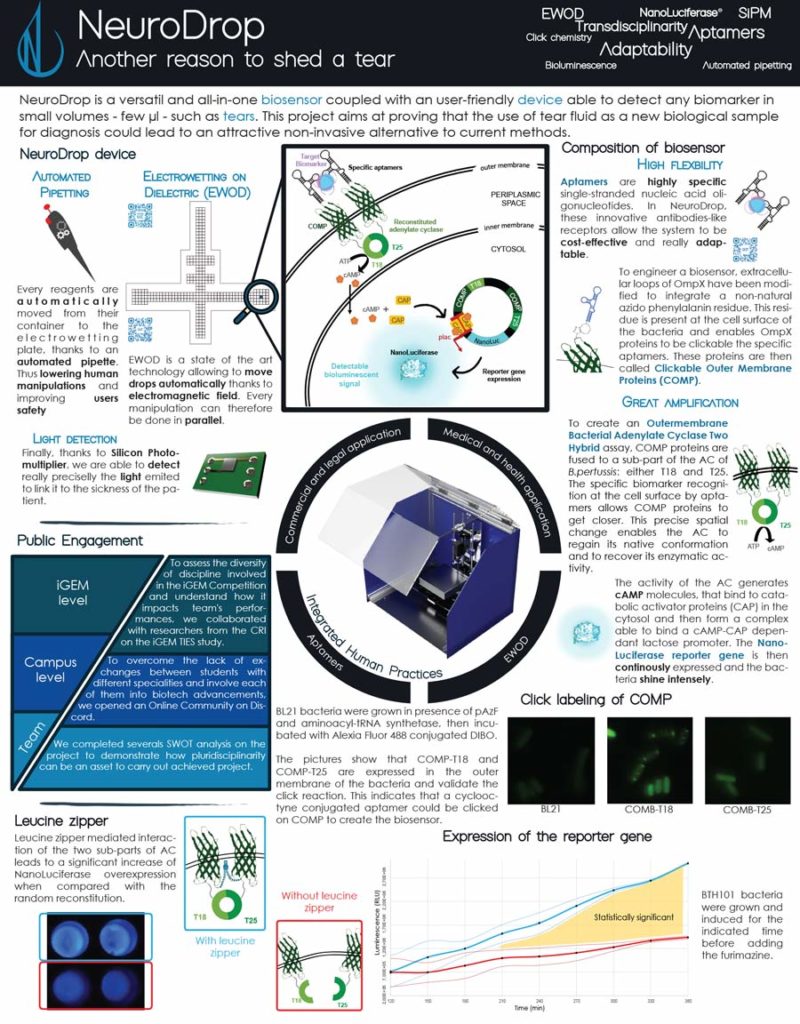
After receiving a NanoLuc® vector from Promega, the team decided to incorporate the bioluminescent reporter into their project. The bright and sensitive signal would be beneficial to their project, but according to team member Lucas Pinero, they ultimately chose NanoLuc® luciferase because it was such a small molecule and didn’t use ATP, which would make it easier for their bacteria to produce the desired proteins.
The team created a biosensor using aptamers (oligonucleotides that bind to specific target molecules) along with Clickable Outer Membrane Proteins (COMP) fused to adenylate cyclase (AC). When an aptamer binds a biomarker, the COMP proteins move closer. The conformation change enables AC activity, producing cAMP molecules that activate a pathway causing the NanoLuc® reporter gene to be continuously expressed and the bacteria to luminesce.
For more details on the Grenoble-Alpes project, which earned a Gold Medal at the 2019 iGEM Giant Jamboree, visit their wiki page.

Easier Enzyme Detection with HiBiT
Sorbonne U Paris decided to focus on developing an alternative to palm oil for their 2019 iGEM project, The Bi[oil]ogical Factory. The most widely used vegetable oil in the world, palm oil production contributes to several negative environmental impacts including deforestation and habitat loss for endangered species. As a proof-of-concept, their project aimed to modify the photosynthetic green microalga Chlamydomonas reinhardtii so it could produce large quantities of palmitic and oleic acid, the main components of palm oil.
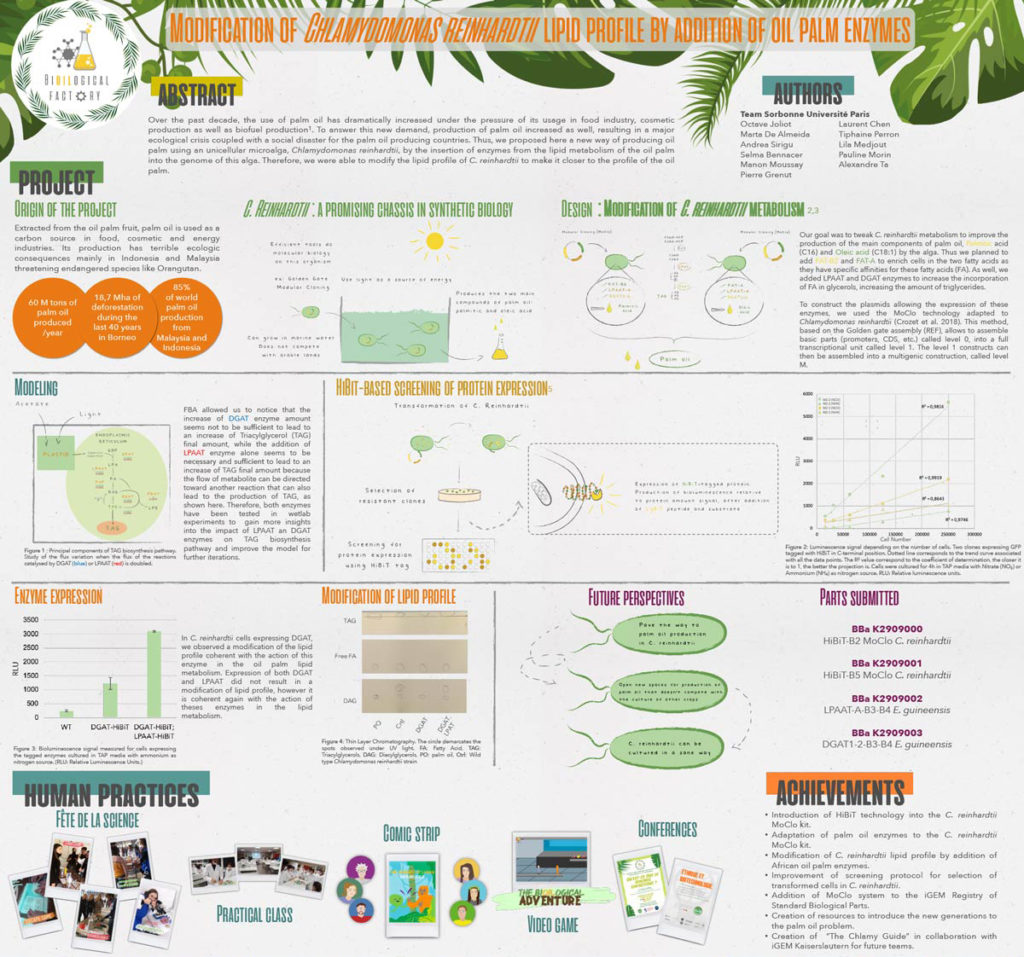
In addition, they decided “to develop a tiny tag for protein detection in C. reinhardtii based on the HiBiT System created by Promega.” One of their team members had worked with HiBiT before and recommended they use it. Their instructor, Pierre Crozet, was familiar with using NanoLuc® luciferase in C. reinhardtii and helped them adapt the standard protocol for HiBiT detection to this organism. The HiBiT tag is the high affinity subunit of the NanoBiT® enzyme. When a HiBiT-tagged protein is incubated with the LgBiT subunit, a functional NanoBiT® enzyme spontaneously assembles and emits a quantifiable luminescence signal in the presence of substrate.
In addition, they decided “to develop a tiny tag for protein detection in C. reinhardtii based on the HiBiT System created by Promega.” One of their team members had worked with HiBiT before and recommended they use it. Their instructor, Pierre Crozet, was familiar with using NanoLuc® luciferase in C. reinhardtii and helped them adapt the standard protocol for HiBiT detection to this organism. The HiBiT tag is basically one part of the NanoBiT® enzyme. When a HiBiT-tagged protein is incubated with the LgBiT (the other part of NanoBiT®) and its substrate, a functional NanoBiT® assembles and emits a quantifiable luminescence signal.
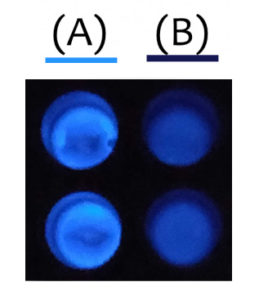
HiBiT allowed them to easily detect and quantify the proteins they wanted to express in C. reinhardtii, the LPAAT-A and DGAT-1-2 enzymes. The HiBiT detection system allowed them to screen lots of clones at the same time for protein expression, precisely quantify protein expression and identify clones with high levels of expression.
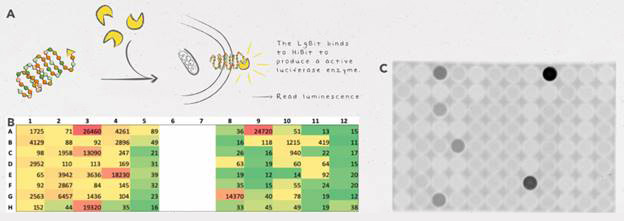
Although the team could have used fluorescent tags, such as GFP, they didn’t want to risk larger tags interfering with the activity of their enzymes. Since HiBiT is so small (11 amino acids in length), it provides easy detection without hindering the function of enzymes as much as other bulkier tags. HiBiT also proved to be a more sensitive method of detection than GFP—although expression levels were too low to detect GFP fluorescence, they were still able to detect protein via the HiBiT tag.
One unexpected observation was the relatively low bioluminescence signal in C. reinhardtii compared with animal cells. Since one of their enzymes was shown to be effective, they hypothesized that the protein was well expressed. After discussing the results with some individuals at Promega and iGEM, they learned that the bioluminescent signal could be quenched by chlorophyll, which was a likely explanation for their results.
Ultimately, the team was able to modify the metabolism and lipid profile of C. reinhardtii by adding genes coding for metabolic enzymes from the oil palm. These results show it is possible to modify C. reinhardtii to have a lipid profile similar to palm oil and increase the levels of lipid production, which could translate to a viable alternative to traditional palm oil production.
Want to see more details about Sorbonne U Paris’s project (which earned a Gold Medal at the 2019 iGEM Giant Jamboree)? Check out their wiki page.

NanoBiT® Reveals Successful Target Binding
MSP-Maastricht’s 2019 iGEM project, RocKit (Receptor Open Community Kit), provides scientists with a tool to create customized cell surface receptors for any target molecule that they choose. By engineering Saccharomyces cerevisiae with the genetic circuitry to produce receptors, they have created a cell than can generate a receptor simply by culturing them in the presence of the target molecule.

They planned to use artificial evolution to induce random mutations at the binding site of a base receptor until it evolved an affinity for the specific target molecule of choice. When the receptor was activated, the mutations would cease and a visible signal would be produced to indicate success.
The team explained what they were trying to accomplish to a Promega representative that visited their lab and said they “received a lot of helpful input for improving their project design.” One critical need was something to tell them when successful target binding had occurred.
The recommended solution was the low affinity NanoBiT® complementation system, consiting of the LgBiT subunit and low affinity 11 amino acid SmBiT. These units can be independently fused to two target proteins to study their interaction. If the two target proteins interact, the SmBiT and LgBiT combine to form the functional NanoBiT® enzyme that produces bright luminescence in the presence of substrate.
This system would signal when the mutations induced in MSP-Maastricht’s engineered organism produced successful target binding as a proof of concept. The NanoBiT® subunits (one on the receptor and one on the target molecule) would come together and interact, producing a very effective visual signal that would tell them target binding had occurred.
Although the team included NanoBiT® in their project design, they ran out of time to complete their experiments and produce any results in time for the Giant Jamboree. One member of the team, Elle Bethune, says she and some teammates “hope to continue the project as their Bachelor’s theses and hope next year’s team will build on the [project] now that all the background research and conceptualization has been done.”
To learn more about MSP-Maastricht’s project, visit their wiki page.

Learn more about how you can get support for your iGEM project at our website.
These three projects represent a small sampling of how this tiny tag can be used in your lab. Click here to learn more about the possibilities for NanoLuc® Technologies in your research.
Related Posts
Latest posts by Darcia Schweitzer (see all)
- Cytochrome P450 Inhibition: Old Drug, New Tricks - May 5, 2022
- Firefly Luciferase Sheds Light on Development of New Malaria Treatments - April 5, 2021
- How to Train Your Instrument Service Team in a Pandemic - February 1, 2021
