One goal of drug discovery and research programs is to reduce false hits as early as possible in the process. Follow-up on false hits is costly in terms of time and resources, and the longer the false hits remain in the drug development pipeline, the more costly they are. So methods that can easily reduce the number of false hits during compound screening early in the discovery process are particularly sought after.
Reporter assays have proven to be invaluable tools for elucidating the mechanisms of action of small molecules or other agents on signaling pathways within cells, and the luciferase reporter assay has become a standard research tool in the biological research laboratory.
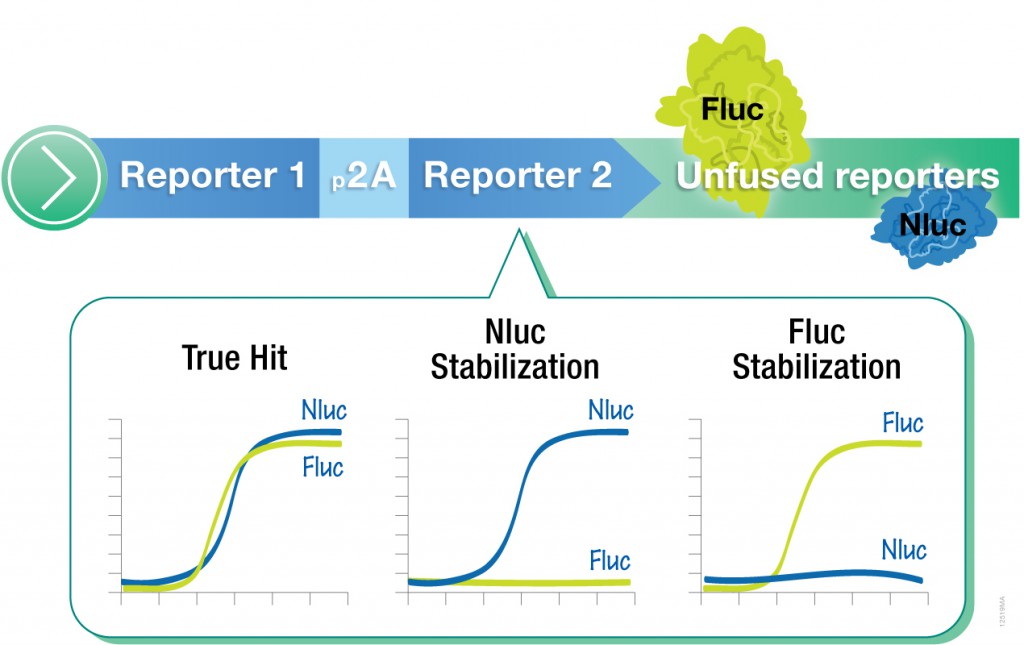
However, one caveat of using standard luciferase-based reporter assays for larger-scale compound screening efforts is the frequency of false hits that result from direct interaction of compounds with the luciferase reporter. This issue can be mitigated with a “coincidence reporter” system where two independent reporter proteins are produced from a single transcript. In this type of assay, a bicistronic transcript is stoichiometrically translated into two nonhomologous reporters by means of a 2A “ribosomal skipping” sequence. Since it is unlikely that compounds will interact with two distinct types of reporter, “coincident” responses will indicate on-target activity. Such a coincident reporter system provides an important control against costly false hits early in drug discovery research programs.
A paper published online in ACS Chem Biol in February describes the first successful application of the firefly/NanoLuc luciferase coincidence reporter system to identify new pathways that up-regulate PARK2 expression.
The PARK2 gene encodes Parkin, an E3 ubiquitin ligase that plays a key role in mitochondrial quality control. When mitochondria are damaged, Parkin mediates a selective autophagic mechanism to facilitate mitochondrial turnover. Loss-of-function mutations in PARK2 are associated with early onset Parkinson’s disease (PD), while genetic enhancement of Parkin expression increases neuron survival in rodent PD models. Therefore, increasing PARK2 transcription is a potential strategy for PD neuroprotection via Parkin-mediated turnover of damaged mitochondria. In this study, the authors used a luciferase-based quantitative high-throughput screen (qHTS) that interrogated a chemogenomic library for up-regulators of PARK2 transcript.
The authors, Hasson et al., developed a coincident reporter assay with firefly and NanoLuc luciferase reporters to accurately detect compound-mediated increases in endogenous PARK2 expression. Using genome editing, they integrated the coincidence reporter into the PARK2 gene locus of neuroblastoma-derived BE(2)-M17 cells. The Nano-Glo® Dual-Luciferase® Reporter Assay System was used to detect both luciferase readouts in the qHTS format. Out of 7,456 chemogenomic compounds screened, 322 were classified as coincident actives. These 322 compounds included diverse chemical classes including epigenetic agents, drugs controlling cholesterol biosynthesis, and JNK inhibitors. The use of coincidence reporters provided the researchers with an efficient screen that eliminated time wasted on investigating reporter-biased false positives.
Here’s the Paper
Hasson SA, Fogel AI, Wang C, MacArthur R, Guha R, Heman-Ackah S, Martin S, Youle RJ, & Inglese J (2015). Chemogenomic Profiling of Endogenous PARK2 Expression Using a Genome-Edited Coincidence Reporter. ACS chemical biology PMID: 25689131
Latest posts by Promega (see all)
- Beyond Ozempic: The New Frontier of Obesity Research - April 18, 2025
- One Health and H5N1: Promega’s Commitment to Holistic Solutions - April 8, 2025
- Overcoming qPCR Inhibitors: Strategies for Reliable Quantification - March 13, 2025

